
Thermacetogenium phaeum (strain ATCC BAA-254 / DSM 26808 / PB)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Thermoanaerobacterales; Thermoanaerobacteraceae; Thermacetogenium; Thermacetogenium phaeum
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
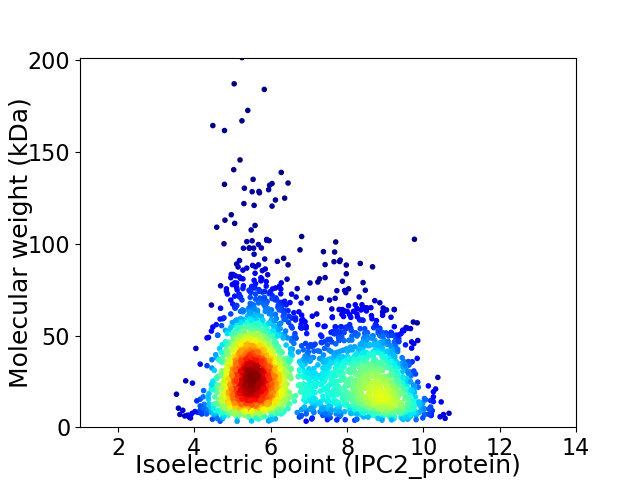
Virtual 2D-PAGE plot for 2803 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4LXA5|K4LXA5_THEPS Transposase OS=Thermacetogenium phaeum (strain ATCC BAA-254 / DSM 26808 / PB) OX=1089553 GN=Tph_c24210 PE=4 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84KK3 pKa = 9.19KK4 pKa = 10.69ALVCQVVVLALALMGFGFAKK24 pKa = 9.99WSDD27 pKa = 3.82SVVVRR32 pKa = 11.84ATAQSGSVKK41 pKa = 9.93WGIFDD46 pKa = 3.54EE47 pKa = 5.17SVWQLDD53 pKa = 4.04EE54 pKa = 4.29GTDD57 pKa = 3.11WHH59 pKa = 8.24SEE61 pKa = 4.09MEE63 pKa = 4.43GGMLNYY69 pKa = 10.72DD70 pKa = 3.37QDD72 pKa = 4.25TEE74 pKa = 4.3GKK76 pKa = 9.77DD77 pKa = 3.3VGSTSVEE84 pKa = 3.89VTDD87 pKa = 4.8ANEE90 pKa = 4.75DD91 pKa = 3.85GVKK94 pKa = 9.54DD95 pKa = 3.7TLDD98 pKa = 3.24ITVSNAYY105 pKa = 8.63PCYY108 pKa = 10.03FNEE111 pKa = 3.87ITFDD115 pKa = 3.81VINAGTIPVIVQIPEE130 pKa = 4.21LTWMEE135 pKa = 4.29TEE137 pKa = 4.47SEE139 pKa = 4.31LEE141 pKa = 4.17SGVVYY146 pKa = 7.81YY147 pKa = 10.79LCSNGSIISEE157 pKa = 4.03YY158 pKa = 10.99DD159 pKa = 3.23DD160 pKa = 5.39GFVSPEE166 pKa = 3.62EE167 pKa = 3.92NGAVIEE173 pKa = 5.73VMFQDD178 pKa = 3.52NVSAQQHH185 pKa = 5.72PGEE188 pKa = 4.36VLSEE192 pKa = 4.2SICFHH197 pKa = 5.95VLQPAAQNDD206 pKa = 3.93QYY208 pKa = 11.73SFSLSIEE215 pKa = 3.83GVQWNEE221 pKa = 3.67SPIPAGSVVDD231 pKa = 3.79
MM1 pKa = 7.7RR2 pKa = 11.84KK3 pKa = 9.19KK4 pKa = 10.69ALVCQVVVLALALMGFGFAKK24 pKa = 9.99WSDD27 pKa = 3.82SVVVRR32 pKa = 11.84ATAQSGSVKK41 pKa = 9.93WGIFDD46 pKa = 3.54EE47 pKa = 5.17SVWQLDD53 pKa = 4.04EE54 pKa = 4.29GTDD57 pKa = 3.11WHH59 pKa = 8.24SEE61 pKa = 4.09MEE63 pKa = 4.43GGMLNYY69 pKa = 10.72DD70 pKa = 3.37QDD72 pKa = 4.25TEE74 pKa = 4.3GKK76 pKa = 9.77DD77 pKa = 3.3VGSTSVEE84 pKa = 3.89VTDD87 pKa = 4.8ANEE90 pKa = 4.75DD91 pKa = 3.85GVKK94 pKa = 9.54DD95 pKa = 3.7TLDD98 pKa = 3.24ITVSNAYY105 pKa = 8.63PCYY108 pKa = 10.03FNEE111 pKa = 3.87ITFDD115 pKa = 3.81VINAGTIPVIVQIPEE130 pKa = 4.21LTWMEE135 pKa = 4.29TEE137 pKa = 4.47SEE139 pKa = 4.31LEE141 pKa = 4.17SGVVYY146 pKa = 7.81YY147 pKa = 10.79LCSNGSIISEE157 pKa = 4.03YY158 pKa = 10.99DD159 pKa = 3.23DD160 pKa = 5.39GFVSPEE166 pKa = 3.62EE167 pKa = 3.92NGAVIEE173 pKa = 5.73VMFQDD178 pKa = 3.52NVSAQQHH185 pKa = 5.72PGEE188 pKa = 4.36VLSEE192 pKa = 4.2SICFHH197 pKa = 5.95VLQPAAQNDD206 pKa = 3.93QYY208 pKa = 11.73SFSLSIEE215 pKa = 3.83GVQWNEE221 pKa = 3.67SPIPAGSVVDD231 pKa = 3.79
Molecular weight: 25.27 kDa
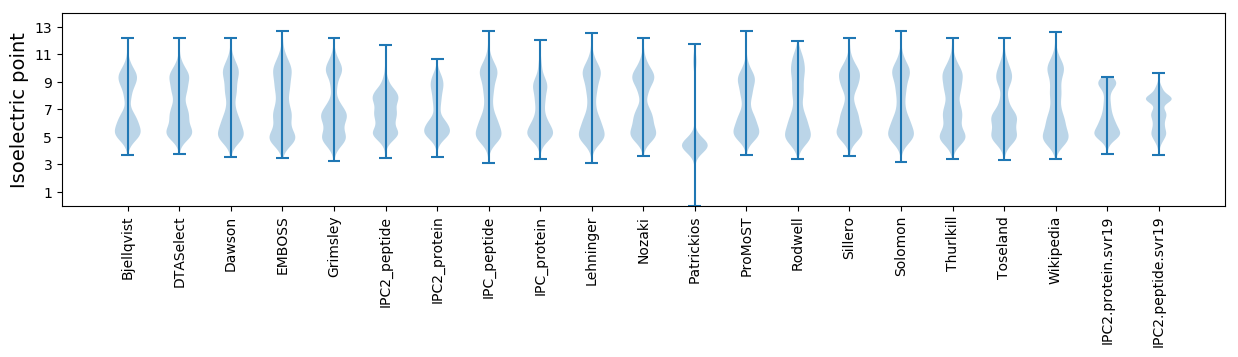
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4LEG3|K4LEG3_THEPS PINc domain-containing protein OS=Thermacetogenium phaeum (strain ATCC BAA-254 / DSM 26808 / PB) OX=1089553 GN=Tph_c01020 PE=4 SV=1
MM1 pKa = 7.21QCGHH5 pKa = 6.92HH6 pKa = 7.3DD7 pKa = 3.29GRR9 pKa = 11.84GPGDD13 pKa = 3.56CLYY16 pKa = 11.13LLRR19 pKa = 11.84HH20 pKa = 5.86QGFEE24 pKa = 4.17GLRR27 pKa = 11.84GTAGEE32 pKa = 3.94QLKK35 pKa = 10.91AGSLHH40 pKa = 6.56RR41 pKa = 11.84ALPRR45 pKa = 11.84PLSGQTPMPPPAAPTEE61 pKa = 4.22YY62 pKa = 10.1EE63 pKa = 3.64NTRR66 pKa = 11.84RR67 pKa = 11.84VCRR70 pKa = 11.84RR71 pKa = 11.84ATYY74 pKa = 9.73GRR76 pKa = 11.84PGNRR80 pKa = 11.84TGEE83 pKa = 3.81ARR85 pKa = 11.84MTGRR89 pKa = 11.84DD90 pKa = 3.37ASTDD94 pKa = 3.42TLRR97 pKa = 11.84VSMEE101 pKa = 4.52WICCEE106 pKa = 4.87AKK108 pKa = 10.43RR109 pKa = 11.84CDD111 pKa = 3.38WQKK114 pKa = 11.07QFATCYY120 pKa = 9.91AASQSCPEE128 pKa = 3.83ASRR131 pKa = 11.84RR132 pKa = 11.84PVRR135 pKa = 11.84PGIGSRR141 pKa = 11.84AVDD144 pKa = 3.6RR145 pKa = 11.84QGMRR149 pKa = 11.84SIFMTGMRR157 pKa = 11.84FPKK160 pKa = 9.94RR161 pKa = 11.84GPGAHH166 pKa = 5.99RR167 pKa = 11.84TPVINLLLQPIGRR180 pKa = 11.84WKK182 pKa = 11.03SNALL186 pKa = 3.44
MM1 pKa = 7.21QCGHH5 pKa = 6.92HH6 pKa = 7.3DD7 pKa = 3.29GRR9 pKa = 11.84GPGDD13 pKa = 3.56CLYY16 pKa = 11.13LLRR19 pKa = 11.84HH20 pKa = 5.86QGFEE24 pKa = 4.17GLRR27 pKa = 11.84GTAGEE32 pKa = 3.94QLKK35 pKa = 10.91AGSLHH40 pKa = 6.56RR41 pKa = 11.84ALPRR45 pKa = 11.84PLSGQTPMPPPAAPTEE61 pKa = 4.22YY62 pKa = 10.1EE63 pKa = 3.64NTRR66 pKa = 11.84RR67 pKa = 11.84VCRR70 pKa = 11.84RR71 pKa = 11.84ATYY74 pKa = 9.73GRR76 pKa = 11.84PGNRR80 pKa = 11.84TGEE83 pKa = 3.81ARR85 pKa = 11.84MTGRR89 pKa = 11.84DD90 pKa = 3.37ASTDD94 pKa = 3.42TLRR97 pKa = 11.84VSMEE101 pKa = 4.52WICCEE106 pKa = 4.87AKK108 pKa = 10.43RR109 pKa = 11.84CDD111 pKa = 3.38WQKK114 pKa = 11.07QFATCYY120 pKa = 9.91AASQSCPEE128 pKa = 3.83ASRR131 pKa = 11.84RR132 pKa = 11.84PVRR135 pKa = 11.84PGIGSRR141 pKa = 11.84AVDD144 pKa = 3.6RR145 pKa = 11.84QGMRR149 pKa = 11.84SIFMTGMRR157 pKa = 11.84FPKK160 pKa = 9.94RR161 pKa = 11.84GPGAHH166 pKa = 5.99RR167 pKa = 11.84TPVINLLLQPIGRR180 pKa = 11.84WKK182 pKa = 11.03SNALL186 pKa = 3.44
Molecular weight: 20.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
812553 |
29 |
1762 |
289.9 |
32.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.844 ± 0.054 | 1.41 ± 0.02 |
4.784 ± 0.033 | 7.669 ± 0.052 |
3.786 ± 0.034 | 8.264 ± 0.045 |
1.687 ± 0.018 | 6.185 ± 0.042 |
5.163 ± 0.047 | 10.869 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.019 | 3.041 ± 0.03 |
4.591 ± 0.032 | 3.099 ± 0.025 |
6.941 ± 0.048 | 5.014 ± 0.031 |
4.515 ± 0.027 | 7.778 ± 0.043 |
1.058 ± 0.021 | 3.075 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
