
Rhodobacteraceae bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; unclassified Rhodobacteraceae
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
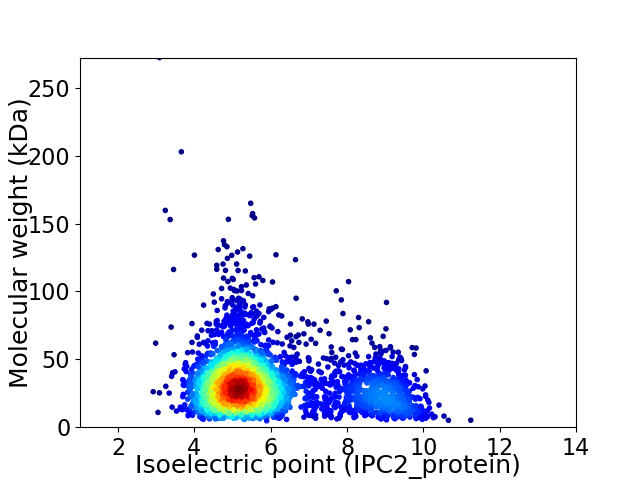
Virtual 2D-PAGE plot for 3201 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
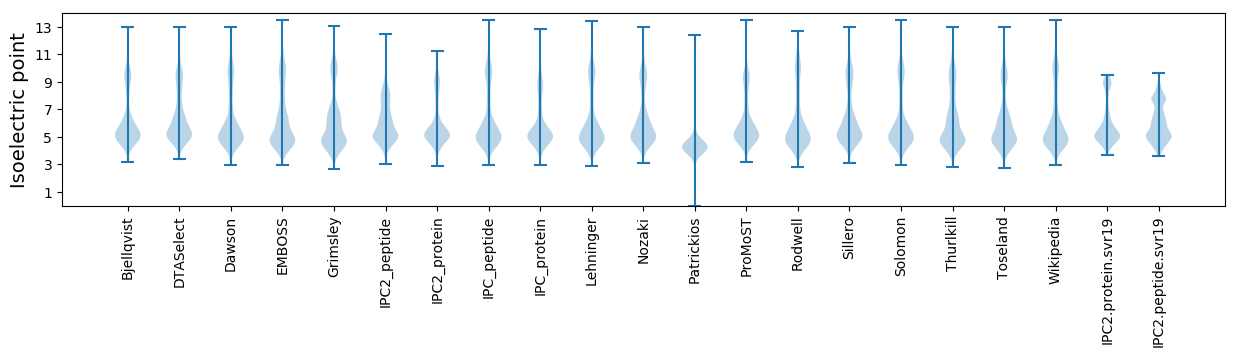
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K9JPW8|A0A2K9JPW8_9RHOB Phosphatidylglycerophosphatase A OS=Rhodobacteraceae bacterium OX=1904441 GN=B9057_06915 PE=4 SV=1
MM1 pKa = 7.27KK2 pKa = 7.86TTLTHH7 pKa = 5.45VAGLCFVATGAIAEE21 pKa = 4.41TSTLTIYY28 pKa = 10.36TYY30 pKa = 11.26DD31 pKa = 3.56GFNSEE36 pKa = 4.84WGPGPQIEE44 pKa = 4.74ANFEE48 pKa = 4.13AEE50 pKa = 4.49CACDD54 pKa = 3.93LEE56 pKa = 5.12FVAAGDD62 pKa = 3.84GAAVLSRR69 pKa = 11.84LKK71 pKa = 11.15LEE73 pKa = 4.68GANTDD78 pKa = 3.14ADD80 pKa = 3.72IVLGLDD86 pKa = 3.41TNLTADD92 pKa = 3.72AAATGLFAASGVTAAYY108 pKa = 8.98DD109 pKa = 4.75LPIEE113 pKa = 4.23WSDD116 pKa = 3.47DD117 pKa = 3.49TFVPFDD123 pKa = 3.01WGYY126 pKa = 9.49FAFVAGKK133 pKa = 10.21DD134 pKa = 3.14VDD136 pKa = 3.61APTNFRR142 pKa = 11.84EE143 pKa = 4.78LAASDD148 pKa = 3.98LKK150 pKa = 10.93IVIQDD155 pKa = 3.85PRR157 pKa = 11.84SSTPGLGLVLWVEE170 pKa = 4.06AAYY173 pKa = 10.44GDD175 pKa = 3.85EE176 pKa = 5.24AGSIWADD183 pKa = 3.18LADD186 pKa = 4.59NIVTVTPGWSEE197 pKa = 4.06AYY199 pKa = 10.34GLFLEE204 pKa = 5.29GEE206 pKa = 4.16ADD208 pKa = 3.5AVLSYY213 pKa = 7.6TTSPAYY219 pKa = 10.26HH220 pKa = 7.29LIAEE224 pKa = 4.75GDD226 pKa = 3.7DD227 pKa = 4.73SKK229 pKa = 10.33TSWAFEE235 pKa = 4.11EE236 pKa = 4.21GHH238 pKa = 5.52YY239 pKa = 9.6MQVEE243 pKa = 4.47VAAKK247 pKa = 9.85VASTDD252 pKa = 3.53VPEE255 pKa = 4.64LADD258 pKa = 3.54AFLAFMTTEE267 pKa = 5.09GFQSVIPTTNWMYY280 pKa = 10.47PAVTPAVGLPEE291 pKa = 4.41GFEE294 pKa = 4.33TLVTPSTALLLSSEE308 pKa = 4.16DD309 pKa = 3.53AAAKK313 pKa = 10.12SGPAIDD319 pKa = 3.62AWRR322 pKa = 11.84AALAQQ327 pKa = 3.73
MM1 pKa = 7.27KK2 pKa = 7.86TTLTHH7 pKa = 5.45VAGLCFVATGAIAEE21 pKa = 4.41TSTLTIYY28 pKa = 10.36TYY30 pKa = 11.26DD31 pKa = 3.56GFNSEE36 pKa = 4.84WGPGPQIEE44 pKa = 4.74ANFEE48 pKa = 4.13AEE50 pKa = 4.49CACDD54 pKa = 3.93LEE56 pKa = 5.12FVAAGDD62 pKa = 3.84GAAVLSRR69 pKa = 11.84LKK71 pKa = 11.15LEE73 pKa = 4.68GANTDD78 pKa = 3.14ADD80 pKa = 3.72IVLGLDD86 pKa = 3.41TNLTADD92 pKa = 3.72AAATGLFAASGVTAAYY108 pKa = 8.98DD109 pKa = 4.75LPIEE113 pKa = 4.23WSDD116 pKa = 3.47DD117 pKa = 3.49TFVPFDD123 pKa = 3.01WGYY126 pKa = 9.49FAFVAGKK133 pKa = 10.21DD134 pKa = 3.14VDD136 pKa = 3.61APTNFRR142 pKa = 11.84EE143 pKa = 4.78LAASDD148 pKa = 3.98LKK150 pKa = 10.93IVIQDD155 pKa = 3.85PRR157 pKa = 11.84SSTPGLGLVLWVEE170 pKa = 4.06AAYY173 pKa = 10.44GDD175 pKa = 3.85EE176 pKa = 5.24AGSIWADD183 pKa = 3.18LADD186 pKa = 4.59NIVTVTPGWSEE197 pKa = 4.06AYY199 pKa = 10.34GLFLEE204 pKa = 5.29GEE206 pKa = 4.16ADD208 pKa = 3.5AVLSYY213 pKa = 7.6TTSPAYY219 pKa = 10.26HH220 pKa = 7.29LIAEE224 pKa = 4.75GDD226 pKa = 3.7DD227 pKa = 4.73SKK229 pKa = 10.33TSWAFEE235 pKa = 4.11EE236 pKa = 4.21GHH238 pKa = 5.52YY239 pKa = 9.6MQVEE243 pKa = 4.47VAAKK247 pKa = 9.85VASTDD252 pKa = 3.53VPEE255 pKa = 4.64LADD258 pKa = 3.54AFLAFMTTEE267 pKa = 5.09GFQSVIPTTNWMYY280 pKa = 10.47PAVTPAVGLPEE291 pKa = 4.41GFEE294 pKa = 4.33TLVTPSTALLLSSEE308 pKa = 4.16DD309 pKa = 3.53AAAKK313 pKa = 10.12SGPAIDD319 pKa = 3.62AWRR322 pKa = 11.84AALAQQ327 pKa = 3.73
Molecular weight: 34.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K9JJ31|A0A2K9JJ31_9RHOB UvrABC system protein B OS=Rhodobacteraceae bacterium OX=1904441 GN=uvrB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
992233 |
41 |
2628 |
310.0 |
33.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.611 ± 0.053 | 0.886 ± 0.013 |
6.322 ± 0.043 | 5.909 ± 0.038 |
3.813 ± 0.025 | 8.413 ± 0.042 |
2.007 ± 0.023 | 5.672 ± 0.03 |
3.569 ± 0.034 | 9.625 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.916 ± 0.023 | 2.943 ± 0.029 |
4.689 ± 0.033 | 3.15 ± 0.023 |
6.203 ± 0.048 | 5.328 ± 0.033 |
5.763 ± 0.038 | 7.483 ± 0.038 |
1.355 ± 0.019 | 2.342 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
