
Corynebacterium sphenisci DSM 44792
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Corynebacteriaceae; Corynebacterium; Corynebacterium sphenisci
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
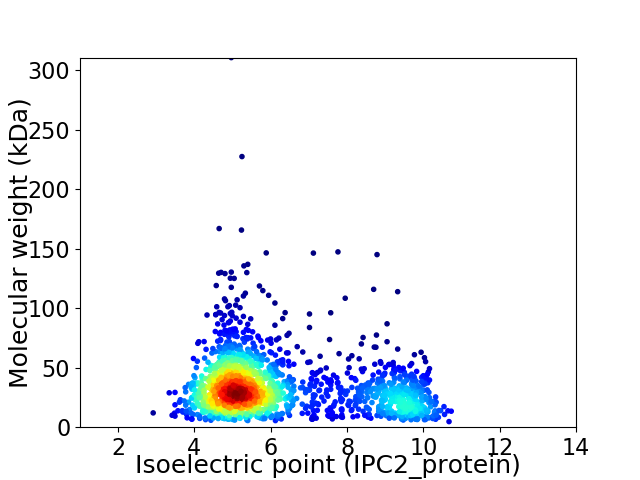
Virtual 2D-PAGE plot for 1827 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
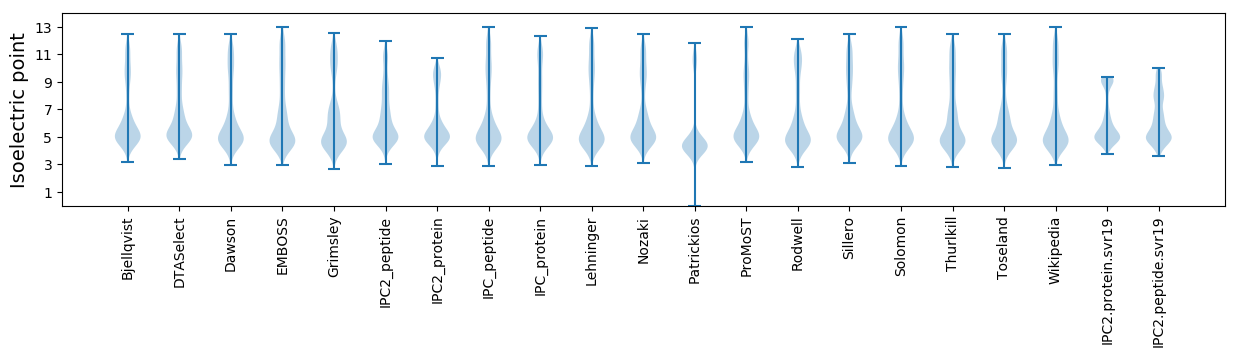
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L7CYY5|A0A1L7CYY5_9CORY ABC transporter OS=Corynebacterium sphenisci DSM 44792 OX=1437874 GN=CSPHI_08825 PE=4 SV=1
MM1 pKa = 7.68NDD3 pKa = 3.31GGFSGDD9 pKa = 3.36ANRR12 pKa = 11.84WGAPGDD18 pKa = 3.76PQGPGPADD26 pKa = 3.66GGAGGPMPEE35 pKa = 4.04QPAWPAEE42 pKa = 4.06NPEE45 pKa = 4.61LTWSADD51 pKa = 3.74PLPPAAPPPAGGHH64 pKa = 6.16PPAGGYY70 pKa = 7.68PPPAGYY76 pKa = 9.39PGAQPAQTAAWSATPHH92 pKa = 6.38AAQLGQQPPPTAMMPPMPAGSTPTGGGAGGGMLPGIAVVAALLLLLGGGGWWWLTQDD149 pKa = 4.77ADD151 pKa = 4.18PAPIAADD158 pKa = 3.27AEE160 pKa = 4.34LAEE163 pKa = 4.72EE164 pKa = 4.9GGFTASAEE172 pKa = 4.07PDD174 pKa = 3.72GEE176 pKa = 4.59DD177 pKa = 3.59GSGAGAEE184 pKa = 4.02AGATGEE190 pKa = 4.41STPPPAPSPTSSRR203 pKa = 11.84STEE206 pKa = 3.73PEE208 pKa = 3.69ARR210 pKa = 11.84SSGCSVDD217 pKa = 4.41ADD219 pKa = 3.56AAVIGRR225 pKa = 11.84AVDD228 pKa = 3.7EE229 pKa = 4.53VNGKK233 pKa = 9.23YY234 pKa = 9.8RR235 pKa = 11.84GRR237 pKa = 11.84WEE239 pKa = 3.74YY240 pKa = 10.75RR241 pKa = 11.84GEE243 pKa = 4.45SNFDD247 pKa = 3.08GCHH250 pKa = 6.11PLTYY254 pKa = 10.45AVLYY258 pKa = 6.57EE259 pKa = 4.16TSGSAGHH266 pKa = 6.34TMLLMFHH273 pKa = 6.63GPDD276 pKa = 3.52YY277 pKa = 11.37LGVDD281 pKa = 3.22SNYY284 pKa = 9.48PQRR287 pKa = 11.84ANSIRR292 pKa = 11.84VTDD295 pKa = 4.14DD296 pKa = 3.22GFIVEE301 pKa = 4.44YY302 pKa = 10.5TDD304 pKa = 5.69LEE306 pKa = 4.77DD307 pKa = 5.07GGDD310 pKa = 3.65SVEE313 pKa = 4.54SVEE316 pKa = 4.14YY317 pKa = 8.99WWDD320 pKa = 3.34GSKK323 pKa = 10.24VAHH326 pKa = 6.77SGRR329 pKa = 11.84IPNTGYY335 pKa = 10.86DD336 pKa = 3.38ALGG339 pKa = 3.48
MM1 pKa = 7.68NDD3 pKa = 3.31GGFSGDD9 pKa = 3.36ANRR12 pKa = 11.84WGAPGDD18 pKa = 3.76PQGPGPADD26 pKa = 3.66GGAGGPMPEE35 pKa = 4.04QPAWPAEE42 pKa = 4.06NPEE45 pKa = 4.61LTWSADD51 pKa = 3.74PLPPAAPPPAGGHH64 pKa = 6.16PPAGGYY70 pKa = 7.68PPPAGYY76 pKa = 9.39PGAQPAQTAAWSATPHH92 pKa = 6.38AAQLGQQPPPTAMMPPMPAGSTPTGGGAGGGMLPGIAVVAALLLLLGGGGWWWLTQDD149 pKa = 4.77ADD151 pKa = 4.18PAPIAADD158 pKa = 3.27AEE160 pKa = 4.34LAEE163 pKa = 4.72EE164 pKa = 4.9GGFTASAEE172 pKa = 4.07PDD174 pKa = 3.72GEE176 pKa = 4.59DD177 pKa = 3.59GSGAGAEE184 pKa = 4.02AGATGEE190 pKa = 4.41STPPPAPSPTSSRR203 pKa = 11.84STEE206 pKa = 3.73PEE208 pKa = 3.69ARR210 pKa = 11.84SSGCSVDD217 pKa = 4.41ADD219 pKa = 3.56AAVIGRR225 pKa = 11.84AVDD228 pKa = 3.7EE229 pKa = 4.53VNGKK233 pKa = 9.23YY234 pKa = 9.8RR235 pKa = 11.84GRR237 pKa = 11.84WEE239 pKa = 3.74YY240 pKa = 10.75RR241 pKa = 11.84GEE243 pKa = 4.45SNFDD247 pKa = 3.08GCHH250 pKa = 6.11PLTYY254 pKa = 10.45AVLYY258 pKa = 6.57EE259 pKa = 4.16TSGSAGHH266 pKa = 6.34TMLLMFHH273 pKa = 6.63GPDD276 pKa = 3.52YY277 pKa = 11.37LGVDD281 pKa = 3.22SNYY284 pKa = 9.48PQRR287 pKa = 11.84ANSIRR292 pKa = 11.84VTDD295 pKa = 4.14DD296 pKa = 3.22GFIVEE301 pKa = 4.44YY302 pKa = 10.5TDD304 pKa = 5.69LEE306 pKa = 4.77DD307 pKa = 5.07GGDD310 pKa = 3.65SVEE313 pKa = 4.54SVEE316 pKa = 4.14YY317 pKa = 8.99WWDD320 pKa = 3.34GSKK323 pKa = 10.24VAHH326 pKa = 6.77SGRR329 pKa = 11.84IPNTGYY335 pKa = 10.86DD336 pKa = 3.38ALGG339 pKa = 3.48
Molecular weight: 34.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L7CVH4|A0A1L7CVH4_9CORY Beta-lactamase OS=Corynebacterium sphenisci DSM 44792 OX=1437874 GN=CSPHI_00795 PE=4 SV=1
MM1 pKa = 7.11NAVAAVWRR9 pKa = 11.84RR10 pKa = 11.84DD11 pKa = 3.36VLRR14 pKa = 11.84TLRR17 pKa = 11.84NKK19 pKa = 10.67AVMVSALLIPGLFLLAFHH37 pKa = 7.15AAFSRR42 pKa = 11.84SAADD46 pKa = 4.24LGIDD50 pKa = 3.56YY51 pKa = 10.9ADD53 pKa = 3.8YY54 pKa = 11.3LLPTAMMQAIIFTAGGSALAVARR77 pKa = 11.84DD78 pKa = 3.89AEE80 pKa = 4.34EE81 pKa = 5.45GIHH84 pKa = 6.33SRR86 pKa = 11.84LRR88 pKa = 11.84TMPAPMWAVAAGRR101 pKa = 11.84VLADD105 pKa = 4.0LTRR108 pKa = 11.84MAWSGGIVLALSLALGARR126 pKa = 11.84IRR128 pKa = 11.84GGIGGILVMLLIFALVTAIIAAGVDD153 pKa = 4.11GLCLLSAKK161 pKa = 9.81PVSTALLFQSLTIIILMFSTAFVPAEE187 pKa = 4.11ALPDD191 pKa = 3.56ALAPVIRR198 pKa = 11.84HH199 pKa = 5.25VPVSPILEE207 pKa = 4.4TIRR210 pKa = 11.84ARR212 pKa = 11.84MAGDD216 pKa = 3.28PAGAAGVEE224 pKa = 3.95AAAWLVALAGAAGWGFVHH242 pKa = 7.09ALSRR246 pKa = 11.84RR247 pKa = 11.84SHH249 pKa = 5.92AA250 pKa = 4.17
MM1 pKa = 7.11NAVAAVWRR9 pKa = 11.84RR10 pKa = 11.84DD11 pKa = 3.36VLRR14 pKa = 11.84TLRR17 pKa = 11.84NKK19 pKa = 10.67AVMVSALLIPGLFLLAFHH37 pKa = 7.15AAFSRR42 pKa = 11.84SAADD46 pKa = 4.24LGIDD50 pKa = 3.56YY51 pKa = 10.9ADD53 pKa = 3.8YY54 pKa = 11.3LLPTAMMQAIIFTAGGSALAVARR77 pKa = 11.84DD78 pKa = 3.89AEE80 pKa = 4.34EE81 pKa = 5.45GIHH84 pKa = 6.33SRR86 pKa = 11.84LRR88 pKa = 11.84TMPAPMWAVAAGRR101 pKa = 11.84VLADD105 pKa = 4.0LTRR108 pKa = 11.84MAWSGGIVLALSLALGARR126 pKa = 11.84IRR128 pKa = 11.84GGIGGILVMLLIFALVTAIIAAGVDD153 pKa = 4.11GLCLLSAKK161 pKa = 9.81PVSTALLFQSLTIIILMFSTAFVPAEE187 pKa = 4.11ALPDD191 pKa = 3.56ALAPVIRR198 pKa = 11.84HH199 pKa = 5.25VPVSPILEE207 pKa = 4.4TIRR210 pKa = 11.84ARR212 pKa = 11.84MAGDD216 pKa = 3.28PAGAAGVEE224 pKa = 3.95AAAWLVALAGAAGWGFVHH242 pKa = 7.09ALSRR246 pKa = 11.84RR247 pKa = 11.84SHH249 pKa = 5.92AA250 pKa = 4.17
Molecular weight: 25.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605766 |
40 |
3043 |
331.6 |
35.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.97 ± 0.122 | 0.725 ± 0.014 |
6.172 ± 0.054 | 6.113 ± 0.065 |
2.667 ± 0.035 | 10.081 ± 0.055 |
2.028 ± 0.025 | 3.937 ± 0.036 |
1.734 ± 0.047 | 9.979 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.988 ± 0.023 | 1.699 ± 0.028 |
6.031 ± 0.054 | 2.086 ± 0.027 |
8.268 ± 0.058 | 3.889 ± 0.037 |
5.125 ± 0.033 | 8.336 ± 0.048 |
1.369 ± 0.025 | 1.803 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
