
Polaribacter sp. MED152
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter; unclassified Polaribacter
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
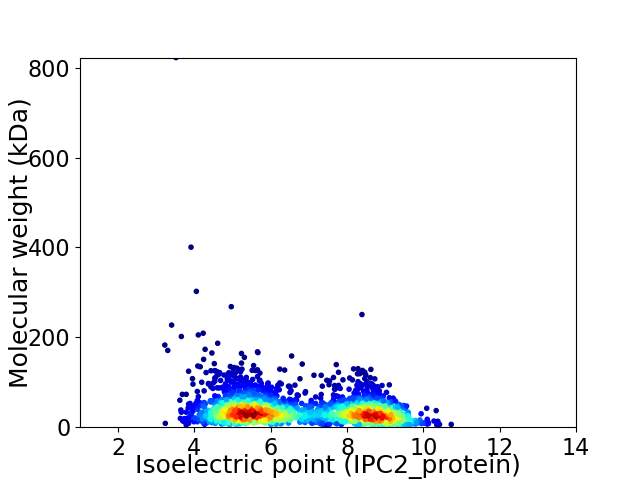
Virtual 2D-PAGE plot for 2634 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
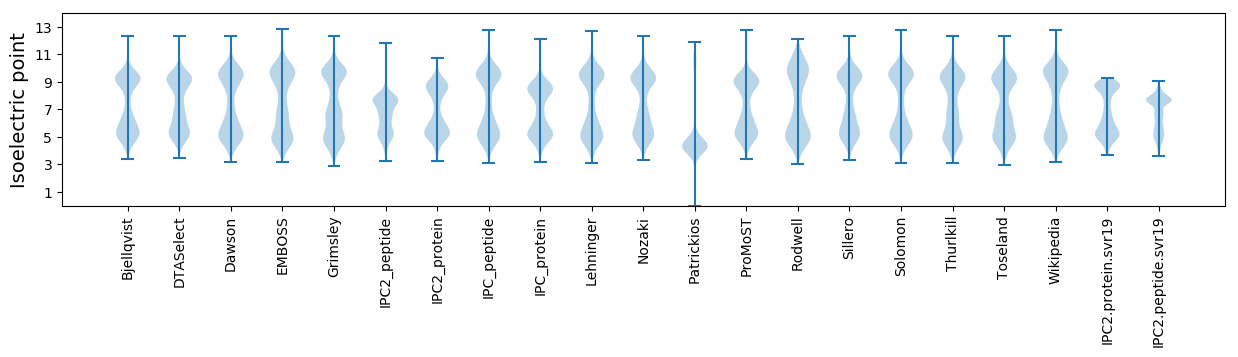
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A2U0Z5|A2U0Z5_9FLAO HI0933-like protein OS=Polaribacter sp. MED152 OX=313598 GN=MED152_04120 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 10.35KK3 pKa = 10.11FLSFLVFLFCASSAFSQLSKK23 pKa = 10.85KK24 pKa = 10.16HH25 pKa = 6.39FIPPLTSVSTDD36 pKa = 3.48EE37 pKa = 5.19IGQQYY42 pKa = 9.87FYY44 pKa = 11.02ISTPKK49 pKa = 9.24TNNVSYY55 pKa = 10.68KK56 pKa = 8.62ITPIGNPDD64 pKa = 2.54ITAYY68 pKa = 10.46SGVVSNGNPFVQPILDD84 pKa = 3.45SSGNFEE90 pKa = 5.46DD91 pKa = 4.55FTSNSQLHH99 pKa = 5.21ITNFNVGGIIKK110 pKa = 10.31NKK112 pKa = 9.95GFIIEE117 pKa = 4.36ASDD120 pKa = 3.6VIYY123 pKa = 11.06VSVRR127 pKa = 11.84VISQSTFHH135 pKa = 6.95AAALVSKK142 pKa = 10.65GSSALGNSFRR152 pKa = 11.84IGGFVRR158 pKa = 11.84EE159 pKa = 4.12GSTVNGHH166 pKa = 5.78LTFASLMATQNNTTVNITNLPAGFTQPNGSNVPNDD201 pKa = 2.92IVLNEE206 pKa = 4.12GEE208 pKa = 4.62SYY210 pKa = 10.91VVALSALAGQDD221 pKa = 3.48PKK223 pKa = 11.65DD224 pKa = 4.28LIGGLISSDD233 pKa = 3.03KK234 pKa = 10.56PIVVNSGSVAGSFGNGNGRR253 pKa = 11.84DD254 pKa = 3.63YY255 pKa = 11.87GMDD258 pKa = 3.46QIVGADD264 pKa = 3.72KK265 pKa = 10.85IGTEE269 pKa = 4.37YY270 pKa = 10.78ILVRR274 pKa = 11.84GSGIDD279 pKa = 3.43SNEE282 pKa = 3.54NALIIAHH289 pKa = 6.47EE290 pKa = 4.48NDD292 pKa = 3.41TEE294 pKa = 4.09ISINGASSTTTIDD307 pKa = 2.87AGEE310 pKa = 4.21YY311 pKa = 10.28FVVEE315 pKa = 4.03GNNYY319 pKa = 10.15NNGNMYY325 pKa = 10.41ISTSKK330 pKa = 10.85AAFVYY335 pKa = 10.31QGIAEE340 pKa = 4.44INNQFPDD347 pKa = 3.49ANQGLFFVPPLSCEE361 pKa = 3.92NKK363 pKa = 10.31GDD365 pKa = 3.76VNNIASIDD373 pKa = 3.95RR374 pKa = 11.84IGSSNFVGGVTIVTNTGATVTINDD398 pKa = 3.8ADD400 pKa = 3.41ISTFSPQGPTNVTGNANYY418 pKa = 8.36VTYY421 pKa = 10.48NVSGLSGNIKK431 pKa = 10.25VNSDD435 pKa = 3.21QEE437 pKa = 4.48LYY439 pKa = 10.31CAYY442 pKa = 10.37FNRR445 pKa = 11.84SGAATTGAFYY455 pKa = 10.77SGFPSAPEE463 pKa = 3.71INFNTTVASLGSCIPNVTLQAANTDD488 pKa = 3.81LFDD491 pKa = 3.82SFEE494 pKa = 4.08WQFFNPATSNWEE506 pKa = 4.01QRR508 pKa = 11.84STTADD513 pKa = 3.55YY514 pKa = 11.04KK515 pKa = 10.72PLEE518 pKa = 4.54SEE520 pKa = 3.77PGRR523 pKa = 11.84YY524 pKa = 9.05RR525 pKa = 11.84LIGTINCTGATFNSVEE541 pKa = 4.46IPVSLCPDD549 pKa = 4.38DD550 pKa = 4.95YY551 pKa = 11.88DD552 pKa = 5.27SDD554 pKa = 5.78LIIDD558 pKa = 4.27NLDD561 pKa = 3.04VDD563 pKa = 5.12LDD565 pKa = 3.77NDD567 pKa = 4.83GILNSDD573 pKa = 3.7EE574 pKa = 4.33SLGDD578 pKa = 3.37GTINLTNLSNPTVEE592 pKa = 4.57ADD594 pKa = 3.55GAIIPNLLTGNLTSDD609 pKa = 3.77GNVSFVGSTNGNFTSTINDD628 pKa = 3.13NSTSNAANIYY638 pKa = 9.21VLNSSEE644 pKa = 4.05TFNFEE649 pKa = 3.74FTQDD653 pKa = 2.72ASQPHH658 pKa = 5.6TSISGVEE665 pKa = 4.0YY666 pKa = 10.43EE667 pKa = 4.62VGIGPAAKK675 pKa = 9.9NVTLIDD681 pKa = 4.14PDD683 pKa = 4.26NILLVDD689 pKa = 3.75TNFDD693 pKa = 3.81GEE695 pKa = 4.39FEE697 pKa = 4.55AGVDD701 pKa = 3.8NYY703 pKa = 10.99SSSLIRR709 pKa = 11.84FTYY712 pKa = 10.47NPAPNGNTPFKK723 pKa = 10.59FVANDD728 pKa = 2.9VDD730 pKa = 4.22EE731 pKa = 5.54VIFLHH736 pKa = 6.1TATSNVTNSIFNGNIKK752 pKa = 8.79LTNVRR757 pKa = 11.84MDD759 pKa = 3.27SDD761 pKa = 3.89GDD763 pKa = 4.18GIEE766 pKa = 4.9DD767 pKa = 5.07ALDD770 pKa = 4.05LDD772 pKa = 4.3SDD774 pKa = 4.08NDD776 pKa = 3.92GVPDD780 pKa = 3.37IFEE783 pKa = 4.51AANEE787 pKa = 4.47SISLLNTDD795 pKa = 4.49ANLDD799 pKa = 3.73GLDD802 pKa = 4.02DD803 pKa = 4.09VFDD806 pKa = 4.23SVTLVNDD813 pKa = 3.52TDD815 pKa = 4.35GDD817 pKa = 4.27GVPNHH822 pKa = 7.19LDD824 pKa = 3.53LDD826 pKa = 4.04SDD828 pKa = 3.75NDD830 pKa = 4.53GIYY833 pKa = 10.95DD834 pKa = 3.77LVEE837 pKa = 4.22SGFTLTDD844 pKa = 3.43ANNDD848 pKa = 4.2GIIDD852 pKa = 3.73NANAANVGINGLLDD866 pKa = 4.06ALEE869 pKa = 4.32STPDD873 pKa = 3.31SGILANALRR882 pKa = 11.84NSDD885 pKa = 3.25ATSVVTANQDD895 pKa = 3.68TIYY898 pKa = 11.29DD899 pKa = 4.01FADD902 pKa = 3.75LDD904 pKa = 4.67ADD906 pKa = 4.26GDD908 pKa = 4.1DD909 pKa = 4.12CFDD912 pKa = 3.86VIEE915 pKa = 5.3AGFTGNGSGILFANPFATDD934 pKa = 3.34ANGLVINNTDD944 pKa = 3.74GYY946 pKa = 6.23TTPNADD952 pKa = 4.54YY953 pKa = 8.9ITSAPIIVNSFVDD966 pKa = 4.15PVFCEE971 pKa = 4.04LNTEE975 pKa = 4.16VMTIDD980 pKa = 3.56SNADD984 pKa = 2.9AFQWQVSTDD993 pKa = 3.49GTNWTSLTDD1002 pKa = 3.65DD1003 pKa = 3.2ATYY1006 pKa = 11.29NGVTTKK1012 pKa = 10.67DD1013 pKa = 3.49LQITDD1018 pKa = 3.85TPLSFNNNQYY1028 pKa = 10.53RR1029 pKa = 11.84VLLSRR1034 pKa = 11.84TGNSCTEE1041 pKa = 4.21EE1042 pKa = 4.13EE1043 pKa = 4.52SSSVTLTVNPLPIIKK1058 pKa = 10.3NNPSEE1063 pKa = 4.1INQCIDD1069 pKa = 3.78ASDD1072 pKa = 3.76TNPTVNLTTSEE1083 pKa = 4.53DD1084 pKa = 4.11NISDD1088 pKa = 3.77TPNITFEE1095 pKa = 4.23YY1096 pKa = 8.8FTDD1099 pKa = 3.49INGTNQITNPTSYY1112 pKa = 10.17PVTVNTIEE1120 pKa = 4.03VVYY1123 pKa = 10.64VSVISDD1129 pKa = 3.65EE1130 pKa = 4.5GCSNGLVEE1138 pKa = 5.36LRR1140 pKa = 11.84INVGQTPDD1148 pKa = 3.21NPYY1151 pKa = 11.1NDD1153 pKa = 3.98IQPPVCDD1160 pKa = 5.08DD1161 pKa = 4.85LLDD1164 pKa = 4.7ADD1166 pKa = 5.41GNDD1169 pKa = 3.65TPGANDD1175 pKa = 3.57DD1176 pKa = 4.04TDD1178 pKa = 4.87FITNFSLDD1186 pKa = 2.84RR1187 pKa = 11.84DD1188 pKa = 4.26AIVNSINPPANTEE1201 pKa = 3.86VFFYY1205 pKa = 10.89EE1206 pKa = 4.52NSSDD1210 pKa = 3.72RR1211 pKa = 11.84NNSLNEE1217 pKa = 3.61IDD1219 pKa = 3.12ITNYY1223 pKa = 10.28RR1224 pKa = 11.84NDD1226 pKa = 3.29INKK1229 pKa = 9.28IDD1231 pKa = 3.65ITTIPEE1237 pKa = 5.2GIQFPIYY1244 pKa = 10.15YY1245 pKa = 10.01KK1246 pKa = 10.46IISTINNNCQGLGEE1260 pKa = 4.67FFLQIHH1266 pKa = 6.72AVPQATTVSDD1276 pKa = 4.01IEE1278 pKa = 4.32EE1279 pKa = 4.57CDD1281 pKa = 3.73DD1282 pKa = 4.07ALSGNTADD1290 pKa = 5.06GRR1292 pKa = 11.84NAAINLRR1299 pKa = 11.84DD1300 pKa = 3.69KK1301 pKa = 10.88VAEE1304 pKa = 3.96ILGTGQTEE1312 pKa = 3.75ADD1314 pKa = 3.89YY1315 pKa = 11.09EE1316 pKa = 4.89VTFHH1320 pKa = 5.4TTEE1323 pKa = 3.65EE1324 pKa = 4.28GAINNIDD1331 pKa = 4.37VITNDD1336 pKa = 3.32TNYY1339 pKa = 10.33TNQAPSGFTAGTTSEE1354 pKa = 3.63QTIYY1358 pKa = 11.06VRR1360 pKa = 11.84VQNRR1364 pKa = 11.84TGSMCYY1370 pKa = 9.78NANTSFKK1377 pKa = 10.45IIIQPIPTVPANVPDD1392 pKa = 4.82LMVCDD1397 pKa = 4.03VVTPSDD1403 pKa = 3.67ADD1405 pKa = 3.33PRR1407 pKa = 11.84NRR1409 pKa = 11.84IAQNINLTEE1418 pKa = 4.15RR1419 pKa = 11.84EE1420 pKa = 3.74ADD1422 pKa = 3.3ILDD1425 pKa = 3.74GRR1427 pKa = 11.84TGLIVEE1433 pKa = 4.76YY1434 pKa = 10.66YY1435 pKa = 10.15ISQQDD1440 pKa = 3.28AEE1442 pKa = 4.29NRR1444 pKa = 11.84TNAIADD1450 pKa = 3.47PSNFQNTTAEE1460 pKa = 4.31TSFPTDD1466 pKa = 3.83FNTDD1470 pKa = 3.46DD1471 pKa = 5.07PGVQTIFFIIVDD1483 pKa = 4.15EE1484 pKa = 5.08NGLQCPSVFSTFQLLVYY1501 pKa = 8.79PEE1503 pKa = 4.32PSINPVSVLSEE1514 pKa = 4.39CDD1516 pKa = 3.36NDD1518 pKa = 5.72DD1519 pKa = 5.75DD1520 pKa = 6.92GDD1522 pKa = 4.05DD1523 pKa = 3.94ANGIIQTIDD1532 pKa = 3.44LSSKK1536 pKa = 9.71IPEE1539 pKa = 3.93VLGASRR1545 pKa = 11.84NVSDD1549 pKa = 4.82FNVTFHH1555 pKa = 7.28ISQADD1560 pKa = 3.49ATSGDD1565 pKa = 3.47NSLASPYY1572 pKa = 9.65TNSNSTEE1579 pKa = 3.97TIYY1582 pKa = 10.61IRR1584 pKa = 11.84VKK1586 pKa = 10.58NKK1588 pKa = 7.58QTMCVNDD1595 pKa = 3.95DD1596 pKa = 3.94ANFEE1600 pKa = 5.04LIVNPLPNFTVTTPQILCLNDD1621 pKa = 3.28LPYY1624 pKa = 10.79NISVEE1629 pKa = 4.25NPSDD1633 pKa = 3.2NYY1635 pKa = 10.09TYY1637 pKa = 11.41VWTDD1641 pKa = 3.16EE1642 pKa = 4.37NGNTLNPNASVDD1654 pKa = 3.96NIDD1657 pKa = 3.61ISSGGNYY1664 pKa = 9.49KK1665 pKa = 8.53VTATTTDD1672 pKa = 2.71GTMCSRR1678 pKa = 11.84EE1679 pKa = 3.82EE1680 pKa = 4.1TIEE1683 pKa = 4.12VIEE1686 pKa = 4.44SNIATLEE1693 pKa = 4.06SSFITIVDD1701 pKa = 3.39EE1702 pKa = 4.5SNNLGSTNNLSISIDD1717 pKa = 3.75TINNDD1722 pKa = 4.53LGPGDD1727 pKa = 3.84YY1728 pKa = 10.69QFAIVNTDD1736 pKa = 2.66NNTRR1740 pKa = 11.84IPFIGYY1746 pKa = 8.49QDD1748 pKa = 3.58EE1749 pKa = 4.36PLFEE1753 pKa = 4.34NLEE1756 pKa = 4.06GGIYY1760 pKa = 9.47QVIVNDD1766 pKa = 4.77KK1767 pKa = 10.06NGCSPDD1773 pKa = 3.3TTLLVSVIQFPKK1785 pKa = 10.42FFTPNGDD1792 pKa = 3.97NINDD1796 pKa = 3.35TWVVKK1801 pKa = 10.13GANQTFYY1808 pKa = 10.51PNASINIFNRR1818 pKa = 11.84FGNLVAQVPIDD1829 pKa = 3.87SNGWNGMYY1837 pKa = 10.04NGKK1840 pKa = 9.23LLPSDD1845 pKa = 4.74DD1846 pKa = 3.13YY1847 pKa = 10.66WYY1849 pKa = 11.35NVTLVPADD1857 pKa = 3.67TTKK1860 pKa = 9.64PTINKK1865 pKa = 9.34KK1866 pKa = 10.95GNFSLLRR1873 pKa = 11.84KK1874 pKa = 9.56
MM1 pKa = 7.56KK2 pKa = 10.35KK3 pKa = 10.11FLSFLVFLFCASSAFSQLSKK23 pKa = 10.85KK24 pKa = 10.16HH25 pKa = 6.39FIPPLTSVSTDD36 pKa = 3.48EE37 pKa = 5.19IGQQYY42 pKa = 9.87FYY44 pKa = 11.02ISTPKK49 pKa = 9.24TNNVSYY55 pKa = 10.68KK56 pKa = 8.62ITPIGNPDD64 pKa = 2.54ITAYY68 pKa = 10.46SGVVSNGNPFVQPILDD84 pKa = 3.45SSGNFEE90 pKa = 5.46DD91 pKa = 4.55FTSNSQLHH99 pKa = 5.21ITNFNVGGIIKK110 pKa = 10.31NKK112 pKa = 9.95GFIIEE117 pKa = 4.36ASDD120 pKa = 3.6VIYY123 pKa = 11.06VSVRR127 pKa = 11.84VISQSTFHH135 pKa = 6.95AAALVSKK142 pKa = 10.65GSSALGNSFRR152 pKa = 11.84IGGFVRR158 pKa = 11.84EE159 pKa = 4.12GSTVNGHH166 pKa = 5.78LTFASLMATQNNTTVNITNLPAGFTQPNGSNVPNDD201 pKa = 2.92IVLNEE206 pKa = 4.12GEE208 pKa = 4.62SYY210 pKa = 10.91VVALSALAGQDD221 pKa = 3.48PKK223 pKa = 11.65DD224 pKa = 4.28LIGGLISSDD233 pKa = 3.03KK234 pKa = 10.56PIVVNSGSVAGSFGNGNGRR253 pKa = 11.84DD254 pKa = 3.63YY255 pKa = 11.87GMDD258 pKa = 3.46QIVGADD264 pKa = 3.72KK265 pKa = 10.85IGTEE269 pKa = 4.37YY270 pKa = 10.78ILVRR274 pKa = 11.84GSGIDD279 pKa = 3.43SNEE282 pKa = 3.54NALIIAHH289 pKa = 6.47EE290 pKa = 4.48NDD292 pKa = 3.41TEE294 pKa = 4.09ISINGASSTTTIDD307 pKa = 2.87AGEE310 pKa = 4.21YY311 pKa = 10.28FVVEE315 pKa = 4.03GNNYY319 pKa = 10.15NNGNMYY325 pKa = 10.41ISTSKK330 pKa = 10.85AAFVYY335 pKa = 10.31QGIAEE340 pKa = 4.44INNQFPDD347 pKa = 3.49ANQGLFFVPPLSCEE361 pKa = 3.92NKK363 pKa = 10.31GDD365 pKa = 3.76VNNIASIDD373 pKa = 3.95RR374 pKa = 11.84IGSSNFVGGVTIVTNTGATVTINDD398 pKa = 3.8ADD400 pKa = 3.41ISTFSPQGPTNVTGNANYY418 pKa = 8.36VTYY421 pKa = 10.48NVSGLSGNIKK431 pKa = 10.25VNSDD435 pKa = 3.21QEE437 pKa = 4.48LYY439 pKa = 10.31CAYY442 pKa = 10.37FNRR445 pKa = 11.84SGAATTGAFYY455 pKa = 10.77SGFPSAPEE463 pKa = 3.71INFNTTVASLGSCIPNVTLQAANTDD488 pKa = 3.81LFDD491 pKa = 3.82SFEE494 pKa = 4.08WQFFNPATSNWEE506 pKa = 4.01QRR508 pKa = 11.84STTADD513 pKa = 3.55YY514 pKa = 11.04KK515 pKa = 10.72PLEE518 pKa = 4.54SEE520 pKa = 3.77PGRR523 pKa = 11.84YY524 pKa = 9.05RR525 pKa = 11.84LIGTINCTGATFNSVEE541 pKa = 4.46IPVSLCPDD549 pKa = 4.38DD550 pKa = 4.95YY551 pKa = 11.88DD552 pKa = 5.27SDD554 pKa = 5.78LIIDD558 pKa = 4.27NLDD561 pKa = 3.04VDD563 pKa = 5.12LDD565 pKa = 3.77NDD567 pKa = 4.83GILNSDD573 pKa = 3.7EE574 pKa = 4.33SLGDD578 pKa = 3.37GTINLTNLSNPTVEE592 pKa = 4.57ADD594 pKa = 3.55GAIIPNLLTGNLTSDD609 pKa = 3.77GNVSFVGSTNGNFTSTINDD628 pKa = 3.13NSTSNAANIYY638 pKa = 9.21VLNSSEE644 pKa = 4.05TFNFEE649 pKa = 3.74FTQDD653 pKa = 2.72ASQPHH658 pKa = 5.6TSISGVEE665 pKa = 4.0YY666 pKa = 10.43EE667 pKa = 4.62VGIGPAAKK675 pKa = 9.9NVTLIDD681 pKa = 4.14PDD683 pKa = 4.26NILLVDD689 pKa = 3.75TNFDD693 pKa = 3.81GEE695 pKa = 4.39FEE697 pKa = 4.55AGVDD701 pKa = 3.8NYY703 pKa = 10.99SSSLIRR709 pKa = 11.84FTYY712 pKa = 10.47NPAPNGNTPFKK723 pKa = 10.59FVANDD728 pKa = 2.9VDD730 pKa = 4.22EE731 pKa = 5.54VIFLHH736 pKa = 6.1TATSNVTNSIFNGNIKK752 pKa = 8.79LTNVRR757 pKa = 11.84MDD759 pKa = 3.27SDD761 pKa = 3.89GDD763 pKa = 4.18GIEE766 pKa = 4.9DD767 pKa = 5.07ALDD770 pKa = 4.05LDD772 pKa = 4.3SDD774 pKa = 4.08NDD776 pKa = 3.92GVPDD780 pKa = 3.37IFEE783 pKa = 4.51AANEE787 pKa = 4.47SISLLNTDD795 pKa = 4.49ANLDD799 pKa = 3.73GLDD802 pKa = 4.02DD803 pKa = 4.09VFDD806 pKa = 4.23SVTLVNDD813 pKa = 3.52TDD815 pKa = 4.35GDD817 pKa = 4.27GVPNHH822 pKa = 7.19LDD824 pKa = 3.53LDD826 pKa = 4.04SDD828 pKa = 3.75NDD830 pKa = 4.53GIYY833 pKa = 10.95DD834 pKa = 3.77LVEE837 pKa = 4.22SGFTLTDD844 pKa = 3.43ANNDD848 pKa = 4.2GIIDD852 pKa = 3.73NANAANVGINGLLDD866 pKa = 4.06ALEE869 pKa = 4.32STPDD873 pKa = 3.31SGILANALRR882 pKa = 11.84NSDD885 pKa = 3.25ATSVVTANQDD895 pKa = 3.68TIYY898 pKa = 11.29DD899 pKa = 4.01FADD902 pKa = 3.75LDD904 pKa = 4.67ADD906 pKa = 4.26GDD908 pKa = 4.1DD909 pKa = 4.12CFDD912 pKa = 3.86VIEE915 pKa = 5.3AGFTGNGSGILFANPFATDD934 pKa = 3.34ANGLVINNTDD944 pKa = 3.74GYY946 pKa = 6.23TTPNADD952 pKa = 4.54YY953 pKa = 8.9ITSAPIIVNSFVDD966 pKa = 4.15PVFCEE971 pKa = 4.04LNTEE975 pKa = 4.16VMTIDD980 pKa = 3.56SNADD984 pKa = 2.9AFQWQVSTDD993 pKa = 3.49GTNWTSLTDD1002 pKa = 3.65DD1003 pKa = 3.2ATYY1006 pKa = 11.29NGVTTKK1012 pKa = 10.67DD1013 pKa = 3.49LQITDD1018 pKa = 3.85TPLSFNNNQYY1028 pKa = 10.53RR1029 pKa = 11.84VLLSRR1034 pKa = 11.84TGNSCTEE1041 pKa = 4.21EE1042 pKa = 4.13EE1043 pKa = 4.52SSSVTLTVNPLPIIKK1058 pKa = 10.3NNPSEE1063 pKa = 4.1INQCIDD1069 pKa = 3.78ASDD1072 pKa = 3.76TNPTVNLTTSEE1083 pKa = 4.53DD1084 pKa = 4.11NISDD1088 pKa = 3.77TPNITFEE1095 pKa = 4.23YY1096 pKa = 8.8FTDD1099 pKa = 3.49INGTNQITNPTSYY1112 pKa = 10.17PVTVNTIEE1120 pKa = 4.03VVYY1123 pKa = 10.64VSVISDD1129 pKa = 3.65EE1130 pKa = 4.5GCSNGLVEE1138 pKa = 5.36LRR1140 pKa = 11.84INVGQTPDD1148 pKa = 3.21NPYY1151 pKa = 11.1NDD1153 pKa = 3.98IQPPVCDD1160 pKa = 5.08DD1161 pKa = 4.85LLDD1164 pKa = 4.7ADD1166 pKa = 5.41GNDD1169 pKa = 3.65TPGANDD1175 pKa = 3.57DD1176 pKa = 4.04TDD1178 pKa = 4.87FITNFSLDD1186 pKa = 2.84RR1187 pKa = 11.84DD1188 pKa = 4.26AIVNSINPPANTEE1201 pKa = 3.86VFFYY1205 pKa = 10.89EE1206 pKa = 4.52NSSDD1210 pKa = 3.72RR1211 pKa = 11.84NNSLNEE1217 pKa = 3.61IDD1219 pKa = 3.12ITNYY1223 pKa = 10.28RR1224 pKa = 11.84NDD1226 pKa = 3.29INKK1229 pKa = 9.28IDD1231 pKa = 3.65ITTIPEE1237 pKa = 5.2GIQFPIYY1244 pKa = 10.15YY1245 pKa = 10.01KK1246 pKa = 10.46IISTINNNCQGLGEE1260 pKa = 4.67FFLQIHH1266 pKa = 6.72AVPQATTVSDD1276 pKa = 4.01IEE1278 pKa = 4.32EE1279 pKa = 4.57CDD1281 pKa = 3.73DD1282 pKa = 4.07ALSGNTADD1290 pKa = 5.06GRR1292 pKa = 11.84NAAINLRR1299 pKa = 11.84DD1300 pKa = 3.69KK1301 pKa = 10.88VAEE1304 pKa = 3.96ILGTGQTEE1312 pKa = 3.75ADD1314 pKa = 3.89YY1315 pKa = 11.09EE1316 pKa = 4.89VTFHH1320 pKa = 5.4TTEE1323 pKa = 3.65EE1324 pKa = 4.28GAINNIDD1331 pKa = 4.37VITNDD1336 pKa = 3.32TNYY1339 pKa = 10.33TNQAPSGFTAGTTSEE1354 pKa = 3.63QTIYY1358 pKa = 11.06VRR1360 pKa = 11.84VQNRR1364 pKa = 11.84TGSMCYY1370 pKa = 9.78NANTSFKK1377 pKa = 10.45IIIQPIPTVPANVPDD1392 pKa = 4.82LMVCDD1397 pKa = 4.03VVTPSDD1403 pKa = 3.67ADD1405 pKa = 3.33PRR1407 pKa = 11.84NRR1409 pKa = 11.84IAQNINLTEE1418 pKa = 4.15RR1419 pKa = 11.84EE1420 pKa = 3.74ADD1422 pKa = 3.3ILDD1425 pKa = 3.74GRR1427 pKa = 11.84TGLIVEE1433 pKa = 4.76YY1434 pKa = 10.66YY1435 pKa = 10.15ISQQDD1440 pKa = 3.28AEE1442 pKa = 4.29NRR1444 pKa = 11.84TNAIADD1450 pKa = 3.47PSNFQNTTAEE1460 pKa = 4.31TSFPTDD1466 pKa = 3.83FNTDD1470 pKa = 3.46DD1471 pKa = 5.07PGVQTIFFIIVDD1483 pKa = 4.15EE1484 pKa = 5.08NGLQCPSVFSTFQLLVYY1501 pKa = 8.79PEE1503 pKa = 4.32PSINPVSVLSEE1514 pKa = 4.39CDD1516 pKa = 3.36NDD1518 pKa = 5.72DD1519 pKa = 5.75DD1520 pKa = 6.92GDD1522 pKa = 4.05DD1523 pKa = 3.94ANGIIQTIDD1532 pKa = 3.44LSSKK1536 pKa = 9.71IPEE1539 pKa = 3.93VLGASRR1545 pKa = 11.84NVSDD1549 pKa = 4.82FNVTFHH1555 pKa = 7.28ISQADD1560 pKa = 3.49ATSGDD1565 pKa = 3.47NSLASPYY1572 pKa = 9.65TNSNSTEE1579 pKa = 3.97TIYY1582 pKa = 10.61IRR1584 pKa = 11.84VKK1586 pKa = 10.58NKK1588 pKa = 7.58QTMCVNDD1595 pKa = 3.95DD1596 pKa = 3.94ANFEE1600 pKa = 5.04LIVNPLPNFTVTTPQILCLNDD1621 pKa = 3.28LPYY1624 pKa = 10.79NISVEE1629 pKa = 4.25NPSDD1633 pKa = 3.2NYY1635 pKa = 10.09TYY1637 pKa = 11.41VWTDD1641 pKa = 3.16EE1642 pKa = 4.37NGNTLNPNASVDD1654 pKa = 3.96NIDD1657 pKa = 3.61ISSGGNYY1664 pKa = 9.49KK1665 pKa = 8.53VTATTTDD1672 pKa = 2.71GTMCSRR1678 pKa = 11.84EE1679 pKa = 3.82EE1680 pKa = 4.1TIEE1683 pKa = 4.12VIEE1686 pKa = 4.44SNIATLEE1693 pKa = 4.06SSFITIVDD1701 pKa = 3.39EE1702 pKa = 4.5SNNLGSTNNLSISIDD1717 pKa = 3.75TINNDD1722 pKa = 4.53LGPGDD1727 pKa = 3.84YY1728 pKa = 10.69QFAIVNTDD1736 pKa = 2.66NNTRR1740 pKa = 11.84IPFIGYY1746 pKa = 8.49QDD1748 pKa = 3.58EE1749 pKa = 4.36PLFEE1753 pKa = 4.34NLEE1756 pKa = 4.06GGIYY1760 pKa = 9.47QVIVNDD1766 pKa = 4.77KK1767 pKa = 10.06NGCSPDD1773 pKa = 3.3TTLLVSVIQFPKK1785 pKa = 10.42FFTPNGDD1792 pKa = 3.97NINDD1796 pKa = 3.35TWVVKK1801 pKa = 10.13GANQTFYY1808 pKa = 10.51PNASINIFNRR1818 pKa = 11.84FGNLVAQVPIDD1829 pKa = 3.87SNGWNGMYY1837 pKa = 10.04NGKK1840 pKa = 9.23LLPSDD1845 pKa = 4.74DD1846 pKa = 3.13YY1847 pKa = 10.66WYY1849 pKa = 11.35NVTLVPADD1857 pKa = 3.67TTKK1860 pKa = 9.64PTINKK1865 pKa = 9.34KK1866 pKa = 10.95GNFSLLRR1873 pKa = 11.84KK1874 pKa = 9.56
Molecular weight: 201.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A2U400|A2U400_9FLAO O-glycosyl hydrolase family 30 OS=Polaribacter sp. MED152 OX=313598 GN=MED152_11364 PE=3 SV=1
MM1 pKa = 6.77TKK3 pKa = 9.03IWKK6 pKa = 9.02FFQYY10 pKa = 10.36GYY12 pKa = 11.32LMVAFICLVEE22 pKa = 4.61GIVRR26 pKa = 11.84FNTEE30 pKa = 3.26RR31 pKa = 11.84TRR33 pKa = 11.84AYY35 pKa = 10.58LFLGFSIFITLVFFFKK51 pKa = 10.37RR52 pKa = 11.84HH53 pKa = 4.19FRR55 pKa = 11.84KK56 pKa = 10.32KK57 pKa = 9.44IEE59 pKa = 4.07KK60 pKa = 10.29RR61 pKa = 11.84NNQNKK66 pKa = 9.6
MM1 pKa = 6.77TKK3 pKa = 9.03IWKK6 pKa = 9.02FFQYY10 pKa = 10.36GYY12 pKa = 11.32LMVAFICLVEE22 pKa = 4.61GIVRR26 pKa = 11.84FNTEE30 pKa = 3.26RR31 pKa = 11.84TRR33 pKa = 11.84AYY35 pKa = 10.58LFLGFSIFITLVFFFKK51 pKa = 10.37RR52 pKa = 11.84HH53 pKa = 4.19FRR55 pKa = 11.84KK56 pKa = 10.32KK57 pKa = 9.44IEE59 pKa = 4.07KK60 pKa = 10.29RR61 pKa = 11.84NNQNKK66 pKa = 9.6
Molecular weight: 8.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
915757 |
35 |
7986 |
347.7 |
39.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.276 ± 0.041 | 0.662 ± 0.017 |
5.636 ± 0.04 | 6.556 ± 0.062 |
5.443 ± 0.047 | 6.12 ± 0.052 |
1.597 ± 0.027 | 8.158 ± 0.048 |
8.358 ± 0.094 | 9.222 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.006 ± 0.03 | 6.779 ± 0.064 |
3.102 ± 0.025 | 3.272 ± 0.025 |
3.17 ± 0.039 | 6.533 ± 0.055 |
5.938 ± 0.105 | 6.235 ± 0.041 |
0.963 ± 0.018 | 3.974 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
