
Mesorhizobium ciceri biovar biserrulae (strain HAMBI 2942 / LMG 23838 / WSM1271)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium; Mesorhizobium ciceri; Mesorhizobium ciceri biovar biserrulae
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
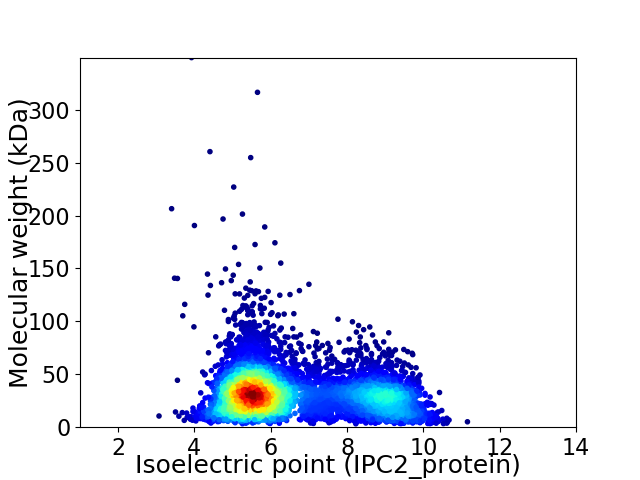
Virtual 2D-PAGE plot for 6260 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E8TH78|E8TH78_MESCW Cobaltochelatase CobN subunit OS=Mesorhizobium ciceri biovar biserrulae (strain HAMBI 2942 / LMG 23838 / WSM1271) OX=765698 GN=Mesci_3280 PE=4 SV=1
MM1 pKa = 7.72LDD3 pKa = 3.07ITAQDD8 pKa = 3.63IIIDD12 pKa = 4.1EE13 pKa = 4.52TTGLQDD19 pKa = 3.26SDD21 pKa = 4.41VNPTVPPHH29 pKa = 6.25NNATVSYY36 pKa = 10.23LLGLDD41 pKa = 3.75GPGGLTSPEE50 pKa = 3.9VAFKK54 pKa = 11.4SNFVVASASAGEE66 pKa = 4.49TITSVVLTQNASGTPFSTTVGVNSGIRR93 pKa = 11.84TVDD96 pKa = 3.25GNYY99 pKa = 9.45VWLFKK104 pKa = 10.81DD105 pKa = 3.18ATHH108 pKa = 6.7ANVVIGVIGTSNPLVAPAATGPLAYY133 pKa = 10.17SFALITTDD141 pKa = 2.91ATHH144 pKa = 7.11ADD146 pKa = 4.67LYY148 pKa = 8.92TVQYY152 pKa = 10.85VPLLHH157 pKa = 6.94PVATDD162 pKa = 3.35PDD164 pKa = 3.97DD165 pKa = 6.32RR166 pKa = 11.84IDD168 pKa = 3.72LTDD171 pKa = 3.55HH172 pKa = 6.13VFASVSGTTVANFSGQNAAPGNHH195 pKa = 6.94DD196 pKa = 4.47FYY198 pKa = 11.36AINSSGGAASQLLVTGFLGANNATANVSTQGFGVNNQSINPTEE241 pKa = 3.96TLQVDD246 pKa = 4.99FVSGANLPAGSASQIQYY263 pKa = 10.54GSHH266 pKa = 6.78IDD268 pKa = 3.98NITHH272 pKa = 7.35AGFTINQITPSNPNLRR288 pKa = 11.84VDD290 pKa = 4.0IKK292 pKa = 10.25ITALDD297 pKa = 3.59VQGNEE302 pKa = 3.95QGLNFYY308 pKa = 10.71DD309 pKa = 4.94GSPTTAAPITSLTLIGQSGVASPITANGTYY339 pKa = 10.43DD340 pKa = 3.13VAGNVDD346 pKa = 3.61VTISGLGTNIVTITGLDD363 pKa = 3.49NITTVNVTTSSQMDD377 pKa = 3.73RR378 pKa = 11.84LLVTGVDD385 pKa = 3.83SNEE388 pKa = 4.03GCDD391 pKa = 2.79ITEE394 pKa = 4.15FHH396 pKa = 7.31FSTTTTNAYY405 pKa = 8.33TEE407 pKa = 4.27QVGSFINFDD416 pKa = 3.77DD417 pKa = 5.43DD418 pKa = 5.13GPTLSITAAPVVGAAEE434 pKa = 4.31VIEE437 pKa = 4.04ASGAGGHH444 pKa = 5.49SQATITPPTFTASAVDD460 pKa = 3.96GVTTNVTYY468 pKa = 11.18ALALAGGAATGLLTTEE484 pKa = 4.34GNHH487 pKa = 7.34AITLVVDD494 pKa = 4.07SANQISGQYY503 pKa = 10.58DD504 pKa = 3.29SDD506 pKa = 3.93GDD508 pKa = 4.23SVLDD512 pKa = 3.47ATAFTVTLSGTTVTLTSLVALEE534 pKa = 4.58HH535 pKa = 7.03SNTQGVGEE543 pKa = 5.13DD544 pKa = 3.35NTLDD548 pKa = 3.78LNGLINVVATVTATDD563 pKa = 3.54GDD565 pKa = 4.04NDD567 pKa = 3.85VVSQQSSSSGLSLTFDD583 pKa = 3.84DD584 pKa = 4.76TDD586 pKa = 3.6PTLSITAATVVGAAEE601 pKa = 4.17VVEE604 pKa = 4.03ASGAGGHH611 pKa = 5.49SQATITPPTFTASAVDD627 pKa = 3.96GVTTNVTYY635 pKa = 11.18ALALAGGAATGLLTTEE651 pKa = 4.34GNHH654 pKa = 7.33AITLVADD661 pKa = 3.84SANQISGQYY670 pKa = 10.58DD671 pKa = 3.29SDD673 pKa = 3.93GDD675 pKa = 4.23SVLDD679 pKa = 3.47ATAFTVTLSGTTVTLTSLVALEE701 pKa = 5.01HH702 pKa = 6.78SNAPQGVGEE711 pKa = 5.5DD712 pKa = 3.39NTLDD716 pKa = 3.78LNGLINVVATVTATDD731 pKa = 3.54GDD733 pKa = 4.04NDD735 pKa = 3.87VVSQQSTSSGLSLTFDD751 pKa = 3.84DD752 pKa = 4.76TDD754 pKa = 3.6PTLSITAATVVGAAGVIEE772 pKa = 3.97ASGVGGQATITPPAFTASAVDD793 pKa = 3.93GVTTNVTYY801 pKa = 11.18ALALAGGAATGLLTTEE817 pKa = 4.34GNHH820 pKa = 7.34AITLVVDD827 pKa = 3.94SATQVSGQYY836 pKa = 10.77DD837 pKa = 3.51SDD839 pKa = 3.93GDD841 pKa = 4.23SVLDD845 pKa = 3.47ATAFTVTLSGTTVTLTSLVALEE867 pKa = 4.58HH868 pKa = 7.03SNTQGVGEE876 pKa = 5.13DD877 pKa = 3.35NTLDD881 pKa = 3.78LNGLINVVATVTATDD896 pKa = 3.54GDD898 pKa = 4.04NDD900 pKa = 3.85VVSQQSSSSGPLSLTFDD917 pKa = 3.96DD918 pKa = 4.45TDD920 pKa = 3.62PTITVPFDD928 pKa = 3.56GDD930 pKa = 3.42QNAGNGTGTHH940 pKa = 4.98EE941 pKa = 4.26TLANTLNASAIGAFGYY957 pKa = 11.26DD958 pKa = 3.09MVDD961 pKa = 2.53KK962 pKa = 9.75HH963 pKa = 5.59TAAEE967 pKa = 4.14YY968 pKa = 10.58LAGASDD974 pKa = 3.92FVDD977 pKa = 3.41VNGALAGIQITLDD990 pKa = 3.48GNLTGLVPAPGTTPFLSSVATLQSEE1015 pKa = 4.82SATSATFNWTASYY1028 pKa = 11.32DD1029 pKa = 3.9SDD1031 pKa = 4.51PNTAGIQPGSVGGTLVFHH1049 pKa = 7.35KK1050 pKa = 10.77DD1051 pKa = 2.65AGTYY1055 pKa = 9.25TITLSDD1061 pKa = 3.61TVEE1064 pKa = 4.52GFTKK1068 pKa = 10.64DD1069 pKa = 3.0ILHH1072 pKa = 6.12TSEE1075 pKa = 5.62LLFKK1079 pKa = 10.7EE1080 pKa = 4.13PLSNTGHH1087 pKa = 6.65PNIVVEE1093 pKa = 4.94KK1094 pKa = 10.4LFEE1097 pKa = 5.22ADD1099 pKa = 3.63STPEE1103 pKa = 3.69TTDD1106 pKa = 2.76RR1107 pKa = 11.84DD1108 pKa = 3.85FFVQFTGNSNPNGSPLGFNATGDD1131 pKa = 3.87GAPAGLPNNLDD1142 pKa = 3.5TAFDD1146 pKa = 3.94AGQQISSNFEE1156 pKa = 3.58DD1157 pKa = 4.59WISATQATNGVAGDD1171 pKa = 4.49TIQKK1175 pKa = 10.86GEE1177 pKa = 4.31LLTLRR1182 pKa = 11.84FFDD1185 pKa = 3.95HH1186 pKa = 6.85SPGIVTDD1193 pKa = 4.04DD1194 pKa = 3.63GVNNVPNQSATDD1206 pKa = 3.61MAIKK1210 pKa = 10.25FDD1212 pKa = 4.67GIGNSEE1218 pKa = 4.65DD1219 pKa = 4.2LMLILNLVNYY1229 pKa = 9.79GSDD1232 pKa = 4.09GIAGGSGTAADD1243 pKa = 4.1TFTTKK1248 pKa = 10.65AMYY1251 pKa = 10.34VSNTNIFKK1259 pKa = 10.76AGQVPTAYY1267 pKa = 10.37AADD1270 pKa = 4.05FPLDD1274 pKa = 3.81NNDD1277 pKa = 3.21GLVIIEE1283 pKa = 4.36KK1284 pKa = 10.26NDD1286 pKa = 3.36YY1287 pKa = 10.47NATGEE1292 pKa = 4.0NWVLQGAQIMQSGNGLTGTAIDD1314 pKa = 4.32LNKK1317 pKa = 9.96ATGTGTTGASTGTHH1331 pKa = 6.11AFDD1334 pKa = 3.57VTDD1337 pKa = 3.78NDD1339 pKa = 3.58VLKK1342 pKa = 9.79ITDD1345 pKa = 3.34IGFTSTQTTTPDD1357 pKa = 2.8AHH1359 pKa = 7.95LDD1361 pKa = 3.53FAFQVADD1368 pKa = 3.59ADD1370 pKa = 4.06GDD1372 pKa = 4.13VTVPQHH1378 pKa = 6.18ILVDD1382 pKa = 4.0VVV1384 pKa = 3.19
MM1 pKa = 7.72LDD3 pKa = 3.07ITAQDD8 pKa = 3.63IIIDD12 pKa = 4.1EE13 pKa = 4.52TTGLQDD19 pKa = 3.26SDD21 pKa = 4.41VNPTVPPHH29 pKa = 6.25NNATVSYY36 pKa = 10.23LLGLDD41 pKa = 3.75GPGGLTSPEE50 pKa = 3.9VAFKK54 pKa = 11.4SNFVVASASAGEE66 pKa = 4.49TITSVVLTQNASGTPFSTTVGVNSGIRR93 pKa = 11.84TVDD96 pKa = 3.25GNYY99 pKa = 9.45VWLFKK104 pKa = 10.81DD105 pKa = 3.18ATHH108 pKa = 6.7ANVVIGVIGTSNPLVAPAATGPLAYY133 pKa = 10.17SFALITTDD141 pKa = 2.91ATHH144 pKa = 7.11ADD146 pKa = 4.67LYY148 pKa = 8.92TVQYY152 pKa = 10.85VPLLHH157 pKa = 6.94PVATDD162 pKa = 3.35PDD164 pKa = 3.97DD165 pKa = 6.32RR166 pKa = 11.84IDD168 pKa = 3.72LTDD171 pKa = 3.55HH172 pKa = 6.13VFASVSGTTVANFSGQNAAPGNHH195 pKa = 6.94DD196 pKa = 4.47FYY198 pKa = 11.36AINSSGGAASQLLVTGFLGANNATANVSTQGFGVNNQSINPTEE241 pKa = 3.96TLQVDD246 pKa = 4.99FVSGANLPAGSASQIQYY263 pKa = 10.54GSHH266 pKa = 6.78IDD268 pKa = 3.98NITHH272 pKa = 7.35AGFTINQITPSNPNLRR288 pKa = 11.84VDD290 pKa = 4.0IKK292 pKa = 10.25ITALDD297 pKa = 3.59VQGNEE302 pKa = 3.95QGLNFYY308 pKa = 10.71DD309 pKa = 4.94GSPTTAAPITSLTLIGQSGVASPITANGTYY339 pKa = 10.43DD340 pKa = 3.13VAGNVDD346 pKa = 3.61VTISGLGTNIVTITGLDD363 pKa = 3.49NITTVNVTTSSQMDD377 pKa = 3.73RR378 pKa = 11.84LLVTGVDD385 pKa = 3.83SNEE388 pKa = 4.03GCDD391 pKa = 2.79ITEE394 pKa = 4.15FHH396 pKa = 7.31FSTTTTNAYY405 pKa = 8.33TEE407 pKa = 4.27QVGSFINFDD416 pKa = 3.77DD417 pKa = 5.43DD418 pKa = 5.13GPTLSITAAPVVGAAEE434 pKa = 4.31VIEE437 pKa = 4.04ASGAGGHH444 pKa = 5.49SQATITPPTFTASAVDD460 pKa = 3.96GVTTNVTYY468 pKa = 11.18ALALAGGAATGLLTTEE484 pKa = 4.34GNHH487 pKa = 7.34AITLVVDD494 pKa = 4.07SANQISGQYY503 pKa = 10.58DD504 pKa = 3.29SDD506 pKa = 3.93GDD508 pKa = 4.23SVLDD512 pKa = 3.47ATAFTVTLSGTTVTLTSLVALEE534 pKa = 4.58HH535 pKa = 7.03SNTQGVGEE543 pKa = 5.13DD544 pKa = 3.35NTLDD548 pKa = 3.78LNGLINVVATVTATDD563 pKa = 3.54GDD565 pKa = 4.04NDD567 pKa = 3.85VVSQQSSSSGLSLTFDD583 pKa = 3.84DD584 pKa = 4.76TDD586 pKa = 3.6PTLSITAATVVGAAEE601 pKa = 4.17VVEE604 pKa = 4.03ASGAGGHH611 pKa = 5.49SQATITPPTFTASAVDD627 pKa = 3.96GVTTNVTYY635 pKa = 11.18ALALAGGAATGLLTTEE651 pKa = 4.34GNHH654 pKa = 7.33AITLVADD661 pKa = 3.84SANQISGQYY670 pKa = 10.58DD671 pKa = 3.29SDD673 pKa = 3.93GDD675 pKa = 4.23SVLDD679 pKa = 3.47ATAFTVTLSGTTVTLTSLVALEE701 pKa = 5.01HH702 pKa = 6.78SNAPQGVGEE711 pKa = 5.5DD712 pKa = 3.39NTLDD716 pKa = 3.78LNGLINVVATVTATDD731 pKa = 3.54GDD733 pKa = 4.04NDD735 pKa = 3.87VVSQQSTSSGLSLTFDD751 pKa = 3.84DD752 pKa = 4.76TDD754 pKa = 3.6PTLSITAATVVGAAGVIEE772 pKa = 3.97ASGVGGQATITPPAFTASAVDD793 pKa = 3.93GVTTNVTYY801 pKa = 11.18ALALAGGAATGLLTTEE817 pKa = 4.34GNHH820 pKa = 7.34AITLVVDD827 pKa = 3.94SATQVSGQYY836 pKa = 10.77DD837 pKa = 3.51SDD839 pKa = 3.93GDD841 pKa = 4.23SVLDD845 pKa = 3.47ATAFTVTLSGTTVTLTSLVALEE867 pKa = 4.58HH868 pKa = 7.03SNTQGVGEE876 pKa = 5.13DD877 pKa = 3.35NTLDD881 pKa = 3.78LNGLINVVATVTATDD896 pKa = 3.54GDD898 pKa = 4.04NDD900 pKa = 3.85VVSQQSSSSGPLSLTFDD917 pKa = 3.96DD918 pKa = 4.45TDD920 pKa = 3.62PTITVPFDD928 pKa = 3.56GDD930 pKa = 3.42QNAGNGTGTHH940 pKa = 4.98EE941 pKa = 4.26TLANTLNASAIGAFGYY957 pKa = 11.26DD958 pKa = 3.09MVDD961 pKa = 2.53KK962 pKa = 9.75HH963 pKa = 5.59TAAEE967 pKa = 4.14YY968 pKa = 10.58LAGASDD974 pKa = 3.92FVDD977 pKa = 3.41VNGALAGIQITLDD990 pKa = 3.48GNLTGLVPAPGTTPFLSSVATLQSEE1015 pKa = 4.82SATSATFNWTASYY1028 pKa = 11.32DD1029 pKa = 3.9SDD1031 pKa = 4.51PNTAGIQPGSVGGTLVFHH1049 pKa = 7.35KK1050 pKa = 10.77DD1051 pKa = 2.65AGTYY1055 pKa = 9.25TITLSDD1061 pKa = 3.61TVEE1064 pKa = 4.52GFTKK1068 pKa = 10.64DD1069 pKa = 3.0ILHH1072 pKa = 6.12TSEE1075 pKa = 5.62LLFKK1079 pKa = 10.7EE1080 pKa = 4.13PLSNTGHH1087 pKa = 6.65PNIVVEE1093 pKa = 4.94KK1094 pKa = 10.4LFEE1097 pKa = 5.22ADD1099 pKa = 3.63STPEE1103 pKa = 3.69TTDD1106 pKa = 2.76RR1107 pKa = 11.84DD1108 pKa = 3.85FFVQFTGNSNPNGSPLGFNATGDD1131 pKa = 3.87GAPAGLPNNLDD1142 pKa = 3.5TAFDD1146 pKa = 3.94AGQQISSNFEE1156 pKa = 3.58DD1157 pKa = 4.59WISATQATNGVAGDD1171 pKa = 4.49TIQKK1175 pKa = 10.86GEE1177 pKa = 4.31LLTLRR1182 pKa = 11.84FFDD1185 pKa = 3.95HH1186 pKa = 6.85SPGIVTDD1193 pKa = 4.04DD1194 pKa = 3.63GVNNVPNQSATDD1206 pKa = 3.61MAIKK1210 pKa = 10.25FDD1212 pKa = 4.67GIGNSEE1218 pKa = 4.65DD1219 pKa = 4.2LMLILNLVNYY1229 pKa = 9.79GSDD1232 pKa = 4.09GIAGGSGTAADD1243 pKa = 4.1TFTTKK1248 pKa = 10.65AMYY1251 pKa = 10.34VSNTNIFKK1259 pKa = 10.76AGQVPTAYY1267 pKa = 10.37AADD1270 pKa = 4.05FPLDD1274 pKa = 3.81NNDD1277 pKa = 3.21GLVIIEE1283 pKa = 4.36KK1284 pKa = 10.26NDD1286 pKa = 3.36YY1287 pKa = 10.47NATGEE1292 pKa = 4.0NWVLQGAQIMQSGNGLTGTAIDD1314 pKa = 4.32LNKK1317 pKa = 9.96ATGTGTTGASTGTHH1331 pKa = 6.11AFDD1334 pKa = 3.57VTDD1337 pKa = 3.78NDD1339 pKa = 3.58VLKK1342 pKa = 9.79ITDD1345 pKa = 3.34IGFTSTQTTTPDD1357 pKa = 2.8AHH1359 pKa = 7.95LDD1361 pKa = 3.53FAFQVADD1368 pKa = 3.59ADD1370 pKa = 4.06GDD1372 pKa = 4.13VTVPQHH1378 pKa = 6.18ILVDD1382 pKa = 4.0VVV1384 pKa = 3.19
Molecular weight: 140.55 kDa
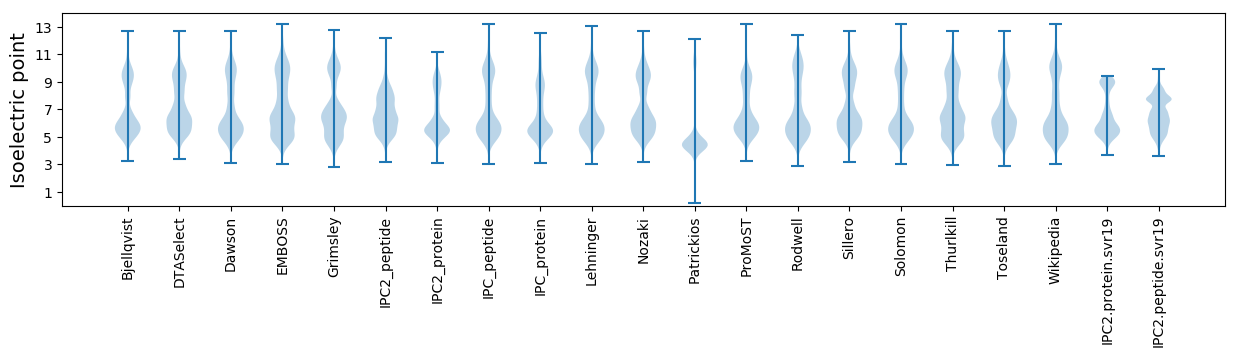
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E8TDS6|E8TDS6_MESCW Sarcosine oxidase delta subunit family protein OS=Mesorhizobium ciceri biovar biserrulae (strain HAMBI 2942 / LMG 23838 / WSM1271) OX=765698 GN=Mesci_1815 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84GVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84GVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1921430 |
30 |
3582 |
306.9 |
33.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.319 ± 0.042 | 0.823 ± 0.01 |
5.709 ± 0.023 | 5.405 ± 0.03 |
3.906 ± 0.02 | 8.706 ± 0.047 |
2.017 ± 0.014 | 5.486 ± 0.021 |
3.703 ± 0.026 | 9.859 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.53 ± 0.014 | 2.751 ± 0.019 |
5.015 ± 0.023 | 3.058 ± 0.018 |
6.658 ± 0.038 | 5.658 ± 0.021 |
5.35 ± 0.028 | 7.463 ± 0.024 |
1.337 ± 0.014 | 2.246 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
