
Wenling tombus-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
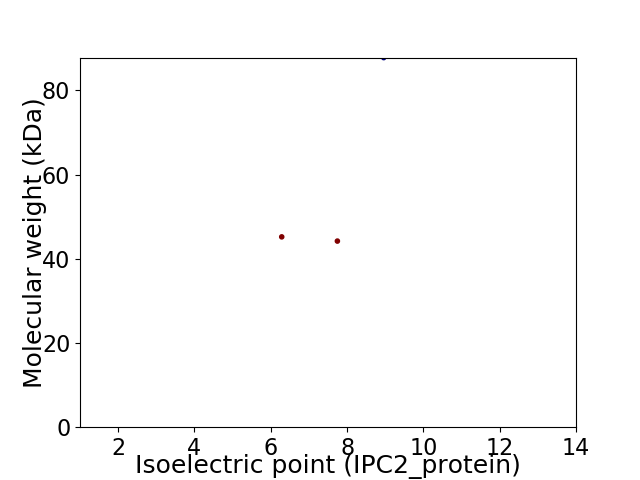
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGT2|A0A1L3KGT2_9VIRU Chropara_Vmeth domain-containing protein OS=Wenling tombus-like virus 5 OX=1923547 PE=4 SV=1
MM1 pKa = 7.75PGAKK5 pKa = 9.44PKK7 pKa = 9.05PQRR10 pKa = 11.84VTRR13 pKa = 11.84TPRR16 pKa = 11.84RR17 pKa = 11.84NAQQRR22 pKa = 11.84SVTQQLQSTRR32 pKa = 11.84QVVNLVASNANHH44 pKa = 6.45RR45 pKa = 11.84PVDD48 pKa = 3.74NSTHH52 pKa = 6.59RR53 pKa = 11.84LNGSDD58 pKa = 3.7YY59 pKa = 10.89LGEE62 pKa = 4.27VTTATPLTAKK72 pKa = 9.17NTVRR76 pKa = 11.84LSMQLSPTGFKK87 pKa = 9.19GTRR90 pKa = 11.84VSALGEE96 pKa = 3.94LYY98 pKa = 10.33EE99 pKa = 4.25KK100 pKa = 10.58YY101 pKa = 9.8RR102 pKa = 11.84YY103 pKa = 9.18RR104 pKa = 11.84RR105 pKa = 11.84AVVRR109 pKa = 11.84YY110 pKa = 9.37VPALPATVGGQLLAYY125 pKa = 9.99VDD127 pKa = 4.5QDD129 pKa = 3.65PKK131 pKa = 11.33DD132 pKa = 3.97DD133 pKa = 3.87PRR135 pKa = 11.84EE136 pKa = 4.05VQDD139 pKa = 4.49LKK141 pKa = 11.28ALRR144 pKa = 11.84AMASSSSGAQAWNLPLAKK162 pKa = 9.92SITMPGTNEE171 pKa = 3.86NKK173 pKa = 9.24WYY175 pKa = 8.46YY176 pKa = 9.9TGHH179 pKa = 5.46QEE181 pKa = 4.03EE182 pKa = 4.41NPRR185 pKa = 11.84LSVQGVLHH193 pKa = 6.95LLQMTDD199 pKa = 3.07AVGTDD204 pKa = 3.94GQPLAANSVAGTLYY218 pKa = 10.73LDD220 pKa = 3.41WVLEE224 pKa = 4.35FKK226 pKa = 9.99TPQMNPEE233 pKa = 4.11LVAAAKK239 pKa = 10.47GYY241 pKa = 10.74VPDD244 pKa = 4.33VPIGPPGGGKK254 pKa = 9.35HH255 pKa = 4.87VTLDD259 pKa = 3.4TRR261 pKa = 11.84KK262 pKa = 10.46LGDD265 pKa = 3.73QASSWIPIPSEE276 pKa = 3.93DD277 pKa = 3.89GFITVRR283 pKa = 11.84NIVGCQPNDD292 pKa = 3.07KK293 pKa = 10.91LMEE296 pKa = 4.64HH297 pKa = 7.04KK298 pKa = 11.15GNGFWIGVYY307 pKa = 8.94WSEE310 pKa = 4.12TGSYY314 pKa = 10.78SDD316 pKa = 5.84DD317 pKa = 3.66PLANPIWRR325 pKa = 11.84VKK327 pKa = 10.26YY328 pKa = 10.15INHH331 pKa = 7.09PLSGDD336 pKa = 2.82MWTSWFYY343 pKa = 11.31LADD346 pKa = 5.25DD347 pKa = 5.11DD348 pKa = 6.09SSMSPFIPPANAKK361 pKa = 9.64YY362 pKa = 8.95VTLIEE367 pKa = 4.41STDD370 pKa = 3.97FPSGSEE376 pKa = 4.26VPVDD380 pKa = 3.78PVDD383 pKa = 3.53TPLYY387 pKa = 7.95TGCRR391 pKa = 11.84VDD393 pKa = 4.91IMWTHH398 pKa = 5.45VKK400 pKa = 10.65EE401 pKa = 4.28EE402 pKa = 3.9TDD404 pKa = 3.54LLVRR408 pKa = 11.84RR409 pKa = 4.71
MM1 pKa = 7.75PGAKK5 pKa = 9.44PKK7 pKa = 9.05PQRR10 pKa = 11.84VTRR13 pKa = 11.84TPRR16 pKa = 11.84RR17 pKa = 11.84NAQQRR22 pKa = 11.84SVTQQLQSTRR32 pKa = 11.84QVVNLVASNANHH44 pKa = 6.45RR45 pKa = 11.84PVDD48 pKa = 3.74NSTHH52 pKa = 6.59RR53 pKa = 11.84LNGSDD58 pKa = 3.7YY59 pKa = 10.89LGEE62 pKa = 4.27VTTATPLTAKK72 pKa = 9.17NTVRR76 pKa = 11.84LSMQLSPTGFKK87 pKa = 9.19GTRR90 pKa = 11.84VSALGEE96 pKa = 3.94LYY98 pKa = 10.33EE99 pKa = 4.25KK100 pKa = 10.58YY101 pKa = 9.8RR102 pKa = 11.84YY103 pKa = 9.18RR104 pKa = 11.84RR105 pKa = 11.84AVVRR109 pKa = 11.84YY110 pKa = 9.37VPALPATVGGQLLAYY125 pKa = 9.99VDD127 pKa = 4.5QDD129 pKa = 3.65PKK131 pKa = 11.33DD132 pKa = 3.97DD133 pKa = 3.87PRR135 pKa = 11.84EE136 pKa = 4.05VQDD139 pKa = 4.49LKK141 pKa = 11.28ALRR144 pKa = 11.84AMASSSSGAQAWNLPLAKK162 pKa = 9.92SITMPGTNEE171 pKa = 3.86NKK173 pKa = 9.24WYY175 pKa = 8.46YY176 pKa = 9.9TGHH179 pKa = 5.46QEE181 pKa = 4.03EE182 pKa = 4.41NPRR185 pKa = 11.84LSVQGVLHH193 pKa = 6.95LLQMTDD199 pKa = 3.07AVGTDD204 pKa = 3.94GQPLAANSVAGTLYY218 pKa = 10.73LDD220 pKa = 3.41WVLEE224 pKa = 4.35FKK226 pKa = 9.99TPQMNPEE233 pKa = 4.11LVAAAKK239 pKa = 10.47GYY241 pKa = 10.74VPDD244 pKa = 4.33VPIGPPGGGKK254 pKa = 9.35HH255 pKa = 4.87VTLDD259 pKa = 3.4TRR261 pKa = 11.84KK262 pKa = 10.46LGDD265 pKa = 3.73QASSWIPIPSEE276 pKa = 3.93DD277 pKa = 3.89GFITVRR283 pKa = 11.84NIVGCQPNDD292 pKa = 3.07KK293 pKa = 10.91LMEE296 pKa = 4.64HH297 pKa = 7.04KK298 pKa = 11.15GNGFWIGVYY307 pKa = 8.94WSEE310 pKa = 4.12TGSYY314 pKa = 10.78SDD316 pKa = 5.84DD317 pKa = 3.66PLANPIWRR325 pKa = 11.84VKK327 pKa = 10.26YY328 pKa = 10.15INHH331 pKa = 7.09PLSGDD336 pKa = 2.82MWTSWFYY343 pKa = 11.31LADD346 pKa = 5.25DD347 pKa = 5.11DD348 pKa = 6.09SSMSPFIPPANAKK361 pKa = 9.64YY362 pKa = 8.95VTLIEE367 pKa = 4.41STDD370 pKa = 3.97FPSGSEE376 pKa = 4.26VPVDD380 pKa = 3.78PVDD383 pKa = 3.53TPLYY387 pKa = 7.95TGCRR391 pKa = 11.84VDD393 pKa = 4.91IMWTHH398 pKa = 5.45VKK400 pKa = 10.65EE401 pKa = 4.28EE402 pKa = 3.9TDD404 pKa = 3.54LLVRR408 pKa = 11.84RR409 pKa = 4.71
Molecular weight: 45.22 kDa
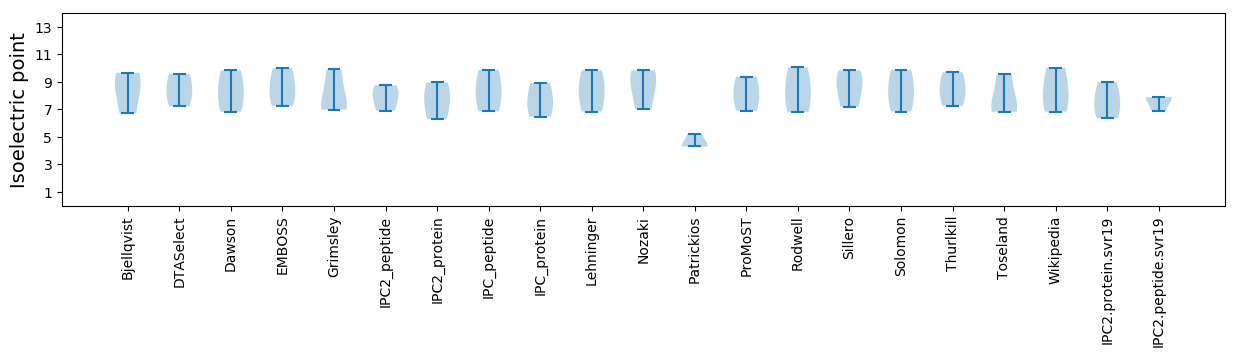
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGZ0|A0A1L3KGZ0_9VIRU RNA-directed RNA polymerase OS=Wenling tombus-like virus 5 OX=1923547 PE=4 SV=1
MM1 pKa = 7.69LPQILLTTLLTMLPGLAPMPEE22 pKa = 3.9LSTYY26 pKa = 9.47EE27 pKa = 3.95WLGLPGLRR35 pKa = 11.84TVRR38 pKa = 11.84HH39 pKa = 5.05VCEE42 pKa = 4.5SSPLPAALHH51 pKa = 5.15WVVSNTLIDD60 pKa = 3.75QDD62 pKa = 3.48RR63 pKa = 11.84LEE65 pKa = 4.5YY66 pKa = 9.71LTAACEE72 pKa = 3.83RR73 pKa = 11.84RR74 pKa = 11.84ADD76 pKa = 3.62ARR78 pKa = 11.84ASLHH82 pKa = 6.58ALTANRR88 pKa = 11.84LWVGTVPEE96 pKa = 4.94LPHH99 pKa = 6.03VCARR103 pKa = 11.84LNSRR107 pKa = 11.84NGYY110 pKa = 9.08IGVPHH115 pKa = 6.42VVIRR119 pKa = 11.84WLEE122 pKa = 3.96SAQLVLATLPSLFLVGGLLSTALFTCIAVAIVRR155 pKa = 11.84YY156 pKa = 9.67RR157 pKa = 11.84CTAHH161 pKa = 6.34VQVACQSPPYY171 pKa = 8.12THH173 pKa = 6.78RR174 pKa = 11.84QVGQMITQRR183 pKa = 11.84LGTWAPCASGDD194 pKa = 3.82NLHH197 pKa = 6.48GHH199 pKa = 6.92LADD202 pKa = 3.66VRR204 pKa = 11.84RR205 pKa = 11.84TLEE208 pKa = 4.04GACFHH213 pKa = 5.85VLKK216 pKa = 10.68SLTGSAGRR224 pKa = 11.84LRR226 pKa = 11.84DD227 pKa = 3.21IGGSLTRR234 pKa = 11.84NARR237 pKa = 11.84LGKK240 pKa = 9.6KK241 pKa = 9.63LHH243 pKa = 6.04VCFPCLTTADD253 pKa = 3.69AARR256 pKa = 11.84LHH258 pKa = 6.39GAQTSNDD265 pKa = 3.74VGHH268 pKa = 6.7HH269 pKa = 6.29TGQDD273 pKa = 3.66CPVSHH278 pKa = 7.15LPSIMTYY285 pKa = 10.64VDD287 pKa = 4.1FHH289 pKa = 5.92MTPSQLVKK297 pKa = 10.47AIKK300 pKa = 10.47SPTLIVTHH308 pKa = 6.83DD309 pKa = 4.09FARR312 pKa = 11.84VGDD315 pKa = 3.88RR316 pKa = 11.84EE317 pKa = 4.3SWFGDD322 pKa = 3.22EE323 pKa = 5.78AIVEE327 pKa = 4.39KK328 pKa = 10.62FGSSVTMTVKK338 pKa = 10.71GGFTYY343 pKa = 9.49THH345 pKa = 7.54GYY347 pKa = 9.28HH348 pKa = 5.77SWGAEE353 pKa = 3.78GTVCTRR359 pKa = 11.84DD360 pKa = 2.91GAFRR364 pKa = 11.84YY365 pKa = 7.62TRR367 pKa = 11.84VYY369 pKa = 10.9DD370 pKa = 3.51NHH372 pKa = 7.2HH373 pKa = 6.74SVILWCCPLAGSFTPSRR390 pKa = 11.84DD391 pKa = 3.39NEE393 pKa = 4.3LVSSVGVVDD402 pKa = 5.45HH403 pKa = 6.5LVCGDD408 pKa = 3.57GTNAAVSGGKK418 pKa = 9.67YY419 pKa = 9.59VFQAGNRR426 pKa = 11.84TLGTVLASTIVRR438 pKa = 11.84VAYY441 pKa = 10.58QMATGARR448 pKa = 11.84DD449 pKa = 3.94EE450 pKa = 4.47KK451 pKa = 10.65WLSNLRR457 pKa = 11.84NLLRR461 pKa = 11.84GRR463 pKa = 11.84FTADD467 pKa = 3.02KK468 pKa = 10.44QDD470 pKa = 3.85EE471 pKa = 4.4NLLPHH476 pKa = 6.77AADD479 pKa = 3.35VCVQLADD486 pKa = 4.34RR487 pKa = 11.84MGTTYY492 pKa = 10.63TNSVLGDD499 pKa = 3.46PSSYY503 pKa = 11.24SFIVRR508 pKa = 11.84PFVRR512 pKa = 11.84YY513 pKa = 8.57AAAAWHH519 pKa = 6.41LVPAPVAACLRR530 pKa = 11.84WATTCVAQVTYY541 pKa = 10.8VRR543 pKa = 11.84SPLTAWAWSTVNVPSYY559 pKa = 10.99EE560 pKa = 4.3VFWDD564 pKa = 3.49QLATVNPSRR573 pKa = 11.84PVPGPFPATGAGSSAAPAQQPHH595 pKa = 6.41SLPGKK600 pKa = 10.06DD601 pKa = 4.22DD602 pKa = 3.89GASGSRR608 pKa = 11.84DD609 pKa = 3.66PEE611 pKa = 4.06KK612 pKa = 10.64SSCGSTPAAPSTGGQPKK629 pKa = 9.0PAAALPPQPLATPPIPLPRR648 pKa = 11.84TTFPAVTPVASKK660 pKa = 10.57SGQDD664 pKa = 3.07SGAAAQAPRR673 pKa = 11.84DD674 pKa = 4.04DD675 pKa = 4.04KK676 pKa = 11.44RR677 pKa = 11.84GVSAGGSKK685 pKa = 8.78GTKK688 pKa = 9.32RR689 pKa = 11.84PVSIPLPPHH698 pKa = 6.1PSEE701 pKa = 4.08TAGARR706 pKa = 11.84LPPKK710 pKa = 10.07ARR712 pKa = 11.84EE713 pKa = 3.92RR714 pKa = 11.84VAKK717 pKa = 10.5GPAANAVRR725 pKa = 11.84PVGVPVQPVKK735 pKa = 10.21TDD737 pKa = 3.35PAQRR741 pKa = 11.84RR742 pKa = 11.84TDD744 pKa = 3.62PSPTTGAIPKK754 pKa = 9.54RR755 pKa = 11.84RR756 pKa = 11.84RR757 pKa = 11.84NEE759 pKa = 3.83GLHH762 pKa = 6.09QDD764 pKa = 3.43GDD766 pKa = 4.05LTKK769 pKa = 11.14GNGSAQHH776 pKa = 5.37QSKK779 pKa = 9.97AAPVSSGSGAPRR791 pKa = 11.84GQPGKK796 pKa = 10.64GSRR799 pKa = 11.84KK800 pKa = 9.6KK801 pKa = 9.93PLSGKK806 pKa = 9.58GANTNRR812 pKa = 11.84KK813 pKa = 8.24GAQAGALPARR823 pKa = 11.84RR824 pKa = 11.84RR825 pKa = 11.84DD826 pKa = 3.34
MM1 pKa = 7.69LPQILLTTLLTMLPGLAPMPEE22 pKa = 3.9LSTYY26 pKa = 9.47EE27 pKa = 3.95WLGLPGLRR35 pKa = 11.84TVRR38 pKa = 11.84HH39 pKa = 5.05VCEE42 pKa = 4.5SSPLPAALHH51 pKa = 5.15WVVSNTLIDD60 pKa = 3.75QDD62 pKa = 3.48RR63 pKa = 11.84LEE65 pKa = 4.5YY66 pKa = 9.71LTAACEE72 pKa = 3.83RR73 pKa = 11.84RR74 pKa = 11.84ADD76 pKa = 3.62ARR78 pKa = 11.84ASLHH82 pKa = 6.58ALTANRR88 pKa = 11.84LWVGTVPEE96 pKa = 4.94LPHH99 pKa = 6.03VCARR103 pKa = 11.84LNSRR107 pKa = 11.84NGYY110 pKa = 9.08IGVPHH115 pKa = 6.42VVIRR119 pKa = 11.84WLEE122 pKa = 3.96SAQLVLATLPSLFLVGGLLSTALFTCIAVAIVRR155 pKa = 11.84YY156 pKa = 9.67RR157 pKa = 11.84CTAHH161 pKa = 6.34VQVACQSPPYY171 pKa = 8.12THH173 pKa = 6.78RR174 pKa = 11.84QVGQMITQRR183 pKa = 11.84LGTWAPCASGDD194 pKa = 3.82NLHH197 pKa = 6.48GHH199 pKa = 6.92LADD202 pKa = 3.66VRR204 pKa = 11.84RR205 pKa = 11.84TLEE208 pKa = 4.04GACFHH213 pKa = 5.85VLKK216 pKa = 10.68SLTGSAGRR224 pKa = 11.84LRR226 pKa = 11.84DD227 pKa = 3.21IGGSLTRR234 pKa = 11.84NARR237 pKa = 11.84LGKK240 pKa = 9.6KK241 pKa = 9.63LHH243 pKa = 6.04VCFPCLTTADD253 pKa = 3.69AARR256 pKa = 11.84LHH258 pKa = 6.39GAQTSNDD265 pKa = 3.74VGHH268 pKa = 6.7HH269 pKa = 6.29TGQDD273 pKa = 3.66CPVSHH278 pKa = 7.15LPSIMTYY285 pKa = 10.64VDD287 pKa = 4.1FHH289 pKa = 5.92MTPSQLVKK297 pKa = 10.47AIKK300 pKa = 10.47SPTLIVTHH308 pKa = 6.83DD309 pKa = 4.09FARR312 pKa = 11.84VGDD315 pKa = 3.88RR316 pKa = 11.84EE317 pKa = 4.3SWFGDD322 pKa = 3.22EE323 pKa = 5.78AIVEE327 pKa = 4.39KK328 pKa = 10.62FGSSVTMTVKK338 pKa = 10.71GGFTYY343 pKa = 9.49THH345 pKa = 7.54GYY347 pKa = 9.28HH348 pKa = 5.77SWGAEE353 pKa = 3.78GTVCTRR359 pKa = 11.84DD360 pKa = 2.91GAFRR364 pKa = 11.84YY365 pKa = 7.62TRR367 pKa = 11.84VYY369 pKa = 10.9DD370 pKa = 3.51NHH372 pKa = 7.2HH373 pKa = 6.74SVILWCCPLAGSFTPSRR390 pKa = 11.84DD391 pKa = 3.39NEE393 pKa = 4.3LVSSVGVVDD402 pKa = 5.45HH403 pKa = 6.5LVCGDD408 pKa = 3.57GTNAAVSGGKK418 pKa = 9.67YY419 pKa = 9.59VFQAGNRR426 pKa = 11.84TLGTVLASTIVRR438 pKa = 11.84VAYY441 pKa = 10.58QMATGARR448 pKa = 11.84DD449 pKa = 3.94EE450 pKa = 4.47KK451 pKa = 10.65WLSNLRR457 pKa = 11.84NLLRR461 pKa = 11.84GRR463 pKa = 11.84FTADD467 pKa = 3.02KK468 pKa = 10.44QDD470 pKa = 3.85EE471 pKa = 4.4NLLPHH476 pKa = 6.77AADD479 pKa = 3.35VCVQLADD486 pKa = 4.34RR487 pKa = 11.84MGTTYY492 pKa = 10.63TNSVLGDD499 pKa = 3.46PSSYY503 pKa = 11.24SFIVRR508 pKa = 11.84PFVRR512 pKa = 11.84YY513 pKa = 8.57AAAAWHH519 pKa = 6.41LVPAPVAACLRR530 pKa = 11.84WATTCVAQVTYY541 pKa = 10.8VRR543 pKa = 11.84SPLTAWAWSTVNVPSYY559 pKa = 10.99EE560 pKa = 4.3VFWDD564 pKa = 3.49QLATVNPSRR573 pKa = 11.84PVPGPFPATGAGSSAAPAQQPHH595 pKa = 6.41SLPGKK600 pKa = 10.06DD601 pKa = 4.22DD602 pKa = 3.89GASGSRR608 pKa = 11.84DD609 pKa = 3.66PEE611 pKa = 4.06KK612 pKa = 10.64SSCGSTPAAPSTGGQPKK629 pKa = 9.0PAAALPPQPLATPPIPLPRR648 pKa = 11.84TTFPAVTPVASKK660 pKa = 10.57SGQDD664 pKa = 3.07SGAAAQAPRR673 pKa = 11.84DD674 pKa = 4.04DD675 pKa = 4.04KK676 pKa = 11.44RR677 pKa = 11.84GVSAGGSKK685 pKa = 8.78GTKK688 pKa = 9.32RR689 pKa = 11.84PVSIPLPPHH698 pKa = 6.1PSEE701 pKa = 4.08TAGARR706 pKa = 11.84LPPKK710 pKa = 10.07ARR712 pKa = 11.84EE713 pKa = 3.92RR714 pKa = 11.84VAKK717 pKa = 10.5GPAANAVRR725 pKa = 11.84PVGVPVQPVKK735 pKa = 10.21TDD737 pKa = 3.35PAQRR741 pKa = 11.84RR742 pKa = 11.84TDD744 pKa = 3.62PSPTTGAIPKK754 pKa = 9.54RR755 pKa = 11.84RR756 pKa = 11.84RR757 pKa = 11.84NEE759 pKa = 3.83GLHH762 pKa = 6.09QDD764 pKa = 3.43GDD766 pKa = 4.05LTKK769 pKa = 11.14GNGSAQHH776 pKa = 5.37QSKK779 pKa = 9.97AAPVSSGSGAPRR791 pKa = 11.84GQPGKK796 pKa = 10.64GSRR799 pKa = 11.84KK800 pKa = 9.6KK801 pKa = 9.93PLSGKK806 pKa = 9.58GANTNRR812 pKa = 11.84KK813 pKa = 8.24GAQAGALPARR823 pKa = 11.84RR824 pKa = 11.84RR825 pKa = 11.84DD826 pKa = 3.34
Molecular weight: 87.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1634 |
399 |
826 |
544.7 |
59.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.792 ± 1.139 | 1.958 ± 0.505 |
4.957 ± 0.502 | 3.305 ± 0.629 |
2.693 ± 0.75 | 7.895 ± 0.781 |
3.06 ± 0.375 | 3.06 ± 0.772 |
3.55 ± 0.299 | 8.813 ± 0.096 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.53 ± 0.344 | 2.938 ± 0.61 |
8.017 ± 0.568 | 3.978 ± 0.26 |
6.916 ± 0.457 | 7.344 ± 0.196 |
7.528 ± 0.3 | 8.262 ± 0.111 |
1.775 ± 0.248 | 2.632 ± 0.467 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
