
Streptococcus phage Javan84
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
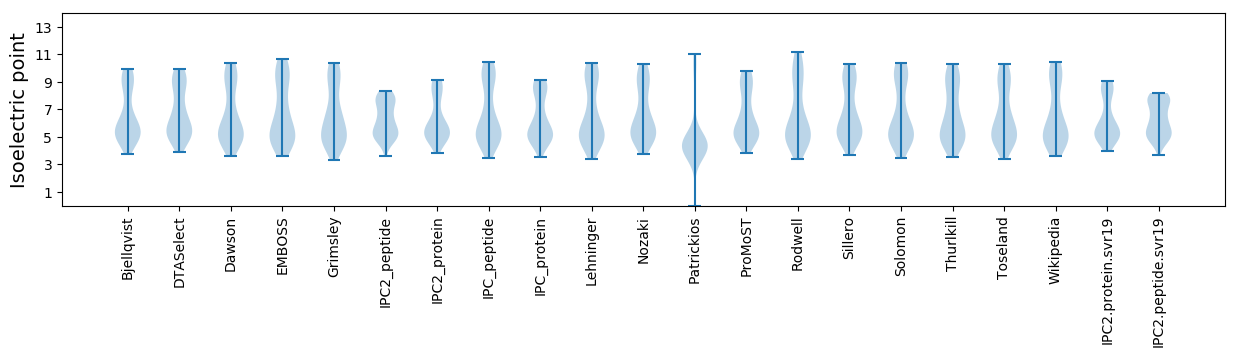
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A4D6BF99|A0A4D6BF99_9CAUD NYN domain-containing protein OS=Streptococcus phage Javan84 OX=2548309 GN=Javan84_0057 PE=4 SV=1
MM1 pKa = 7.72PEE3 pKa = 4.14IYY5 pKa = 10.14QISLVDD11 pKa = 3.65YY12 pKa = 10.9QMEE15 pKa = 4.15LALLVLFDD23 pKa = 4.51YY24 pKa = 10.19YY25 pKa = 10.93TDD27 pKa = 3.58NVLVSSPHH35 pKa = 6.13PLSVVV40 pKa = 3.06
MM1 pKa = 7.72PEE3 pKa = 4.14IYY5 pKa = 10.14QISLVDD11 pKa = 3.65YY12 pKa = 10.9QMEE15 pKa = 4.15LALLVLFDD23 pKa = 4.51YY24 pKa = 10.19YY25 pKa = 10.93TDD27 pKa = 3.58NVLVSSPHH35 pKa = 6.13PLSVVV40 pKa = 3.06
Molecular weight: 4.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6BEQ9|A0A4D6BEQ9_9CAUD Uncharacterized protein OS=Streptococcus phage Javan84 OX=2548309 GN=Javan84_0003 PE=4 SV=1
MM1 pKa = 7.48ASYY4 pKa = 10.53RR5 pKa = 11.84KK6 pKa = 9.31RR7 pKa = 11.84DD8 pKa = 3.51GSWEE12 pKa = 3.63YY13 pKa = 10.59RR14 pKa = 11.84ISYY17 pKa = 7.39KK18 pKa = 10.53TPDD21 pKa = 3.22GKK23 pKa = 11.03YY24 pKa = 8.55KK25 pKa = 10.48QKK27 pKa = 10.61SKK29 pKa = 10.98RR30 pKa = 11.84GFRR33 pKa = 11.84TKK35 pKa = 10.63KK36 pKa = 9.27EE37 pKa = 4.06AEE39 pKa = 3.95LAAAEE44 pKa = 4.39AEE46 pKa = 4.0RR47 pKa = 11.84LLNSTTIFDD56 pKa = 4.39EE57 pKa = 4.74NISLYY62 pKa = 10.79DD63 pKa = 4.67YY64 pKa = 10.37FDD66 pKa = 3.4KK67 pKa = 10.42WAKK70 pKa = 9.78IYY72 pKa = 10.1KK73 pKa = 9.15KK74 pKa = 8.83PHH76 pKa = 5.04VSIGTWKK83 pKa = 10.91NYY85 pKa = 10.35DD86 pKa = 3.36QTLKK90 pKa = 10.88LIEE93 pKa = 4.36RR94 pKa = 11.84EE95 pKa = 3.89FGQTKK100 pKa = 9.46IRR102 pKa = 11.84AITPSIYY109 pKa = 10.09QQFLNNLGKK118 pKa = 10.2RR119 pKa = 11.84YY120 pKa = 9.34YY121 pKa = 10.17QGTIHH126 pKa = 6.85KK127 pKa = 8.32VHH129 pKa = 5.99HH130 pKa = 6.73RR131 pKa = 11.84IRR133 pKa = 11.84RR134 pKa = 11.84AVKK137 pKa = 8.6QAVVEE142 pKa = 4.23KK143 pKa = 10.91LIDD146 pKa = 3.59TNFTDD151 pKa = 3.88LAKK154 pKa = 10.52INAEE158 pKa = 4.0RR159 pKa = 11.84QHH161 pKa = 7.63KK162 pKa = 9.66PIEE165 pKa = 4.44DD166 pKa = 3.4KK167 pKa = 10.89FLEE170 pKa = 4.31EE171 pKa = 4.49NEE173 pKa = 4.16YY174 pKa = 11.22LSLLTTLKK182 pKa = 10.53QNCPQTDD189 pKa = 3.87YY190 pKa = 11.32VQLYY194 pKa = 8.69LLSVTGMRR202 pKa = 11.84IGEE205 pKa = 4.3SLGLTWNDD213 pKa = 2.33IDD215 pKa = 5.06FKK217 pKa = 11.5NGLININKK225 pKa = 7.28TWNVYY230 pKa = 8.84TNAEE234 pKa = 3.98FAPTKK239 pKa = 10.13NKK241 pKa = 10.26QSMRR245 pKa = 11.84TIPLDD250 pKa = 3.22NTTAKK255 pKa = 10.33ILLQFKK261 pKa = 8.92TEE263 pKa = 3.74EE264 pKa = 4.18WTVNPYY270 pKa = 10.07NRR272 pKa = 11.84LFIRR276 pKa = 11.84VNHH279 pKa = 6.34PWLNRR284 pKa = 11.84LIKK287 pKa = 10.55KK288 pKa = 7.59LTKK291 pKa = 10.26TNIHH295 pKa = 4.95VHH297 pKa = 5.1SLRR300 pKa = 11.84HH301 pKa = 5.46TYY303 pKa = 10.81ASYY306 pKa = 10.88LISRR310 pKa = 11.84QIDD313 pKa = 3.81LLTISNLLGHH323 pKa = 7.16KK324 pKa = 10.21DD325 pKa = 3.46LTVTLQTYY333 pKa = 9.12AHH335 pKa = 6.13QLEE338 pKa = 4.39QQKK341 pKa = 10.67EE342 pKa = 3.95KK343 pKa = 11.06DD344 pKa = 3.35FKK346 pKa = 10.85EE347 pKa = 3.89IKK349 pKa = 10.28KK350 pKa = 10.66LFGG353 pKa = 3.74
MM1 pKa = 7.48ASYY4 pKa = 10.53RR5 pKa = 11.84KK6 pKa = 9.31RR7 pKa = 11.84DD8 pKa = 3.51GSWEE12 pKa = 3.63YY13 pKa = 10.59RR14 pKa = 11.84ISYY17 pKa = 7.39KK18 pKa = 10.53TPDD21 pKa = 3.22GKK23 pKa = 11.03YY24 pKa = 8.55KK25 pKa = 10.48QKK27 pKa = 10.61SKK29 pKa = 10.98RR30 pKa = 11.84GFRR33 pKa = 11.84TKK35 pKa = 10.63KK36 pKa = 9.27EE37 pKa = 4.06AEE39 pKa = 3.95LAAAEE44 pKa = 4.39AEE46 pKa = 4.0RR47 pKa = 11.84LLNSTTIFDD56 pKa = 4.39EE57 pKa = 4.74NISLYY62 pKa = 10.79DD63 pKa = 4.67YY64 pKa = 10.37FDD66 pKa = 3.4KK67 pKa = 10.42WAKK70 pKa = 9.78IYY72 pKa = 10.1KK73 pKa = 9.15KK74 pKa = 8.83PHH76 pKa = 5.04VSIGTWKK83 pKa = 10.91NYY85 pKa = 10.35DD86 pKa = 3.36QTLKK90 pKa = 10.88LIEE93 pKa = 4.36RR94 pKa = 11.84EE95 pKa = 3.89FGQTKK100 pKa = 9.46IRR102 pKa = 11.84AITPSIYY109 pKa = 10.09QQFLNNLGKK118 pKa = 10.2RR119 pKa = 11.84YY120 pKa = 9.34YY121 pKa = 10.17QGTIHH126 pKa = 6.85KK127 pKa = 8.32VHH129 pKa = 5.99HH130 pKa = 6.73RR131 pKa = 11.84IRR133 pKa = 11.84RR134 pKa = 11.84AVKK137 pKa = 8.6QAVVEE142 pKa = 4.23KK143 pKa = 10.91LIDD146 pKa = 3.59TNFTDD151 pKa = 3.88LAKK154 pKa = 10.52INAEE158 pKa = 4.0RR159 pKa = 11.84QHH161 pKa = 7.63KK162 pKa = 9.66PIEE165 pKa = 4.44DD166 pKa = 3.4KK167 pKa = 10.89FLEE170 pKa = 4.31EE171 pKa = 4.49NEE173 pKa = 4.16YY174 pKa = 11.22LSLLTTLKK182 pKa = 10.53QNCPQTDD189 pKa = 3.87YY190 pKa = 11.32VQLYY194 pKa = 8.69LLSVTGMRR202 pKa = 11.84IGEE205 pKa = 4.3SLGLTWNDD213 pKa = 2.33IDD215 pKa = 5.06FKK217 pKa = 11.5NGLININKK225 pKa = 7.28TWNVYY230 pKa = 8.84TNAEE234 pKa = 3.98FAPTKK239 pKa = 10.13NKK241 pKa = 10.26QSMRR245 pKa = 11.84TIPLDD250 pKa = 3.22NTTAKK255 pKa = 10.33ILLQFKK261 pKa = 8.92TEE263 pKa = 3.74EE264 pKa = 4.18WTVNPYY270 pKa = 10.07NRR272 pKa = 11.84LFIRR276 pKa = 11.84VNHH279 pKa = 6.34PWLNRR284 pKa = 11.84LIKK287 pKa = 10.55KK288 pKa = 7.59LTKK291 pKa = 10.26TNIHH295 pKa = 4.95VHH297 pKa = 5.1SLRR300 pKa = 11.84HH301 pKa = 5.46TYY303 pKa = 10.81ASYY306 pKa = 10.88LISRR310 pKa = 11.84QIDD313 pKa = 3.81LLTISNLLGHH323 pKa = 7.16KK324 pKa = 10.21DD325 pKa = 3.46LTVTLQTYY333 pKa = 9.12AHH335 pKa = 6.13QLEE338 pKa = 4.39QQKK341 pKa = 10.67EE342 pKa = 3.95KK343 pKa = 11.06DD344 pKa = 3.35FKK346 pKa = 10.85EE347 pKa = 3.89IKK349 pKa = 10.28KK350 pKa = 10.66LFGG353 pKa = 3.74
Molecular weight: 41.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13199 |
40 |
1271 |
227.6 |
25.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.97 ± 0.756 | 0.568 ± 0.102 |
6.076 ± 0.351 | 7.16 ± 0.549 |
3.781 ± 0.178 | 6.447 ± 0.402 |
1.614 ± 0.152 | 6.985 ± 0.279 |
7.448 ± 0.491 | 8.129 ± 0.299 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.705 ± 0.208 | 4.909 ± 0.318 |
3.068 ± 0.201 | 4.25 ± 0.296 |
4.493 ± 0.258 | 6.372 ± 0.454 |
6.213 ± 0.35 | 6.788 ± 0.396 |
1.197 ± 0.165 | 3.826 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
