
Reichenbachiella faecimaris
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Reichenbachiellaceae; Reichenbachiella
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
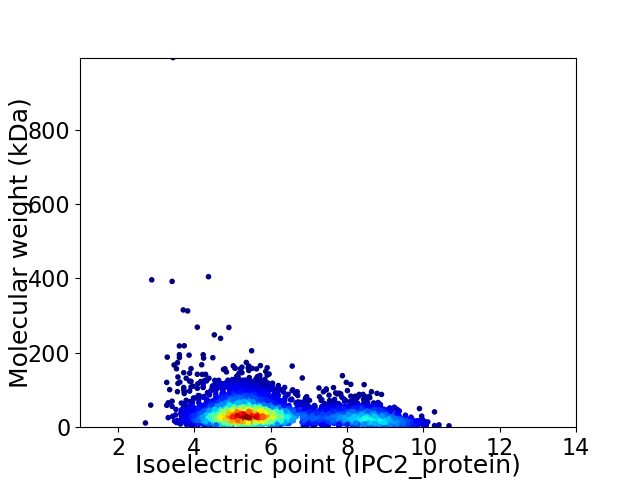
Virtual 2D-PAGE plot for 3972 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W2GHE7|A0A1W2GHE7_9BACT Antitoxin component YwqK of the YwqJK toxin-antitoxin module OS=Reichenbachiella faecimaris OX=692418 GN=SAMN04488029_2702 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 10.33NFKK5 pKa = 10.1IYY7 pKa = 9.87TLILVSACIAFSCDD21 pKa = 3.51EE22 pKa = 4.22EE23 pKa = 5.65DD24 pKa = 5.54KK25 pKa = 11.34LITDD29 pKa = 4.56ILTEE33 pKa = 4.2NPLPVPTGDD42 pKa = 3.9PGEE45 pKa = 5.02LDD47 pKa = 3.33LSTYY51 pKa = 9.86VAIGNSLTAGFQDD64 pKa = 3.1NALYY68 pKa = 10.29TDD70 pKa = 3.8GQINAFPAYY79 pKa = 9.0LTTQFSAQGVGGGDD93 pKa = 3.39YY94 pKa = 10.66VYY96 pKa = 11.0PDD98 pKa = 3.31INSEE102 pKa = 4.0NGYY105 pKa = 10.49SGTDD109 pKa = 3.33GTNLFGKK116 pKa = 10.3YY117 pKa = 9.34ILNITEE123 pKa = 4.2LGIEE127 pKa = 4.25ISAGEE132 pKa = 3.99NNIGLRR138 pKa = 11.84ATEE141 pKa = 4.53DD142 pKa = 3.57EE143 pKa = 5.11IEE145 pKa = 4.12ALTNFGVPGATISDD159 pKa = 3.99LDD161 pKa = 4.1DD162 pKa = 4.37ASFANPYY169 pKa = 9.91FLRR172 pKa = 11.84FSSNPLNVSILDD184 pKa = 3.61DD185 pKa = 3.72VVARR189 pKa = 11.84NPTFISLWIGSNDD202 pKa = 3.4YY203 pKa = 10.94LGYY206 pKa = 10.99ALGGAASGAPTDD218 pKa = 3.52VATFQTMFTNHH229 pKa = 4.57MTKK232 pKa = 10.59LAGTGAKK239 pKa = 9.88GVVLNLPPVTLIPYY253 pKa = 9.21FRR255 pKa = 11.84AVPYY259 pKa = 10.44NAVPLTDD266 pKa = 3.64QSDD269 pKa = 3.82VDD271 pKa = 4.39DD272 pKa = 5.54LNAATAFGGYY282 pKa = 8.75NAALDD287 pKa = 5.17GIAAMDD293 pKa = 4.32PNFSQEE299 pKa = 4.1DD300 pKa = 3.43ADD302 pKa = 4.03EE303 pKa = 4.81RR304 pKa = 11.84KK305 pKa = 8.33VTYY308 pKa = 10.51SLGANPVLIKK318 pKa = 10.54DD319 pKa = 3.67TDD321 pKa = 4.11LVDD324 pKa = 4.02LTTTLAGINPALALYY339 pKa = 9.5GQTRR343 pKa = 11.84QANEE347 pKa = 3.66NDD349 pKa = 4.91LILLPASTLIGTDD362 pKa = 3.39AEE364 pKa = 4.47GLPDD368 pKa = 3.88VPIGVANPLGDD379 pKa = 3.78EE380 pKa = 4.3YY381 pKa = 11.85VLTFSEE387 pKa = 4.33QVEE390 pKa = 4.94VITARR395 pKa = 11.84ATFNGVISAVLANSAFSNFKK415 pKa = 10.65LVDD418 pKa = 3.51IQPFFADD425 pKa = 3.64AFGLSAAQATALALTSDD442 pKa = 5.13AIAAADD448 pKa = 3.65GVLGIEE454 pKa = 4.31YY455 pKa = 10.31QGFDD459 pKa = 3.81YY460 pKa = 11.37SPDD463 pKa = 3.5FTPNGLWSNDD473 pKa = 2.91GVHH476 pKa = 6.63PNPRR480 pKa = 11.84GHH482 pKa = 7.25AIVANEE488 pKa = 4.21IIEE491 pKa = 4.43TMNEE495 pKa = 3.41TWSADD500 pKa = 3.14IPTVTVNQKK509 pKa = 9.59RR510 pKa = 11.84ASPFTQQ516 pKa = 3.08
MM1 pKa = 7.67KK2 pKa = 10.33NFKK5 pKa = 10.1IYY7 pKa = 9.87TLILVSACIAFSCDD21 pKa = 3.51EE22 pKa = 4.22EE23 pKa = 5.65DD24 pKa = 5.54KK25 pKa = 11.34LITDD29 pKa = 4.56ILTEE33 pKa = 4.2NPLPVPTGDD42 pKa = 3.9PGEE45 pKa = 5.02LDD47 pKa = 3.33LSTYY51 pKa = 9.86VAIGNSLTAGFQDD64 pKa = 3.1NALYY68 pKa = 10.29TDD70 pKa = 3.8GQINAFPAYY79 pKa = 9.0LTTQFSAQGVGGGDD93 pKa = 3.39YY94 pKa = 10.66VYY96 pKa = 11.0PDD98 pKa = 3.31INSEE102 pKa = 4.0NGYY105 pKa = 10.49SGTDD109 pKa = 3.33GTNLFGKK116 pKa = 10.3YY117 pKa = 9.34ILNITEE123 pKa = 4.2LGIEE127 pKa = 4.25ISAGEE132 pKa = 3.99NNIGLRR138 pKa = 11.84ATEE141 pKa = 4.53DD142 pKa = 3.57EE143 pKa = 5.11IEE145 pKa = 4.12ALTNFGVPGATISDD159 pKa = 3.99LDD161 pKa = 4.1DD162 pKa = 4.37ASFANPYY169 pKa = 9.91FLRR172 pKa = 11.84FSSNPLNVSILDD184 pKa = 3.61DD185 pKa = 3.72VVARR189 pKa = 11.84NPTFISLWIGSNDD202 pKa = 3.4YY203 pKa = 10.94LGYY206 pKa = 10.99ALGGAASGAPTDD218 pKa = 3.52VATFQTMFTNHH229 pKa = 4.57MTKK232 pKa = 10.59LAGTGAKK239 pKa = 9.88GVVLNLPPVTLIPYY253 pKa = 9.21FRR255 pKa = 11.84AVPYY259 pKa = 10.44NAVPLTDD266 pKa = 3.64QSDD269 pKa = 3.82VDD271 pKa = 4.39DD272 pKa = 5.54LNAATAFGGYY282 pKa = 8.75NAALDD287 pKa = 5.17GIAAMDD293 pKa = 4.32PNFSQEE299 pKa = 4.1DD300 pKa = 3.43ADD302 pKa = 4.03EE303 pKa = 4.81RR304 pKa = 11.84KK305 pKa = 8.33VTYY308 pKa = 10.51SLGANPVLIKK318 pKa = 10.54DD319 pKa = 3.67TDD321 pKa = 4.11LVDD324 pKa = 4.02LTTTLAGINPALALYY339 pKa = 9.5GQTRR343 pKa = 11.84QANEE347 pKa = 3.66NDD349 pKa = 4.91LILLPASTLIGTDD362 pKa = 3.39AEE364 pKa = 4.47GLPDD368 pKa = 3.88VPIGVANPLGDD379 pKa = 3.78EE380 pKa = 4.3YY381 pKa = 11.85VLTFSEE387 pKa = 4.33QVEE390 pKa = 4.94VITARR395 pKa = 11.84ATFNGVISAVLANSAFSNFKK415 pKa = 10.65LVDD418 pKa = 3.51IQPFFADD425 pKa = 3.64AFGLSAAQATALALTSDD442 pKa = 5.13AIAAADD448 pKa = 3.65GVLGIEE454 pKa = 4.31YY455 pKa = 10.31QGFDD459 pKa = 3.81YY460 pKa = 11.37SPDD463 pKa = 3.5FTPNGLWSNDD473 pKa = 2.91GVHH476 pKa = 6.63PNPRR480 pKa = 11.84GHH482 pKa = 7.25AIVANEE488 pKa = 4.21IIEE491 pKa = 4.43TMNEE495 pKa = 3.41TWSADD500 pKa = 3.14IPTVTVNQKK509 pKa = 9.59RR510 pKa = 11.84ASPFTQQ516 pKa = 3.08
Molecular weight: 54.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W2GMQ2|A0A1W2GMQ2_9BACT 3-oxoacyl-[acyl-carrier-protein] reductase OS=Reichenbachiella faecimaris OX=692418 GN=SAMN04488029_3551 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.5GIQEE6 pKa = 4.23FFEE9 pKa = 4.66KK10 pKa = 10.83YY11 pKa = 10.66AFGVCTRR18 pKa = 11.84LGEE21 pKa = 4.23KK22 pKa = 10.34LRR24 pKa = 11.84IPSSSIRR31 pKa = 11.84LFFIYY36 pKa = 10.14TSFLTFGSPVLVYY49 pKa = 11.01LMLAFVMNFRR59 pKa = 11.84KK60 pKa = 10.04HH61 pKa = 3.6IRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84HH66 pKa = 4.71SLWYY70 pKa = 9.8HH71 pKa = 6.01
MM1 pKa = 7.57KK2 pKa = 10.5GIQEE6 pKa = 4.23FFEE9 pKa = 4.66KK10 pKa = 10.83YY11 pKa = 10.66AFGVCTRR18 pKa = 11.84LGEE21 pKa = 4.23KK22 pKa = 10.34LRR24 pKa = 11.84IPSSSIRR31 pKa = 11.84LFFIYY36 pKa = 10.14TSFLTFGSPVLVYY49 pKa = 11.01LMLAFVMNFRR59 pKa = 11.84KK60 pKa = 10.04HH61 pKa = 3.6IRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84HH66 pKa = 4.71SLWYY70 pKa = 9.8HH71 pKa = 6.01
Molecular weight: 8.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1432065 |
29 |
9392 |
360.5 |
40.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.695 ± 0.039 | 0.749 ± 0.012 |
5.946 ± 0.053 | 6.835 ± 0.04 |
5.001 ± 0.029 | 6.782 ± 0.045 |
1.919 ± 0.02 | 7.42 ± 0.035 |
6.747 ± 0.06 | 9.453 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.416 ± 0.024 | 5.456 ± 0.034 |
3.35 ± 0.021 | 3.636 ± 0.023 |
3.593 ± 0.03 | 6.99 ± 0.035 |
5.519 ± 0.055 | 6.328 ± 0.031 |
1.146 ± 0.016 | 4.02 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
