
Flavonifractor sp. An135
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Flavonifractor; unclassified Flavonifractor
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
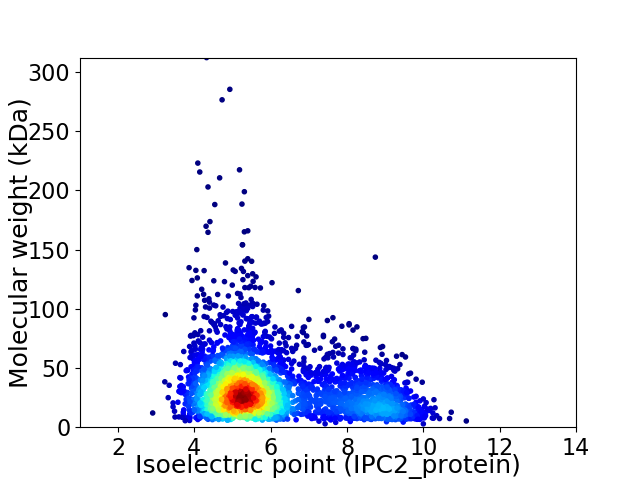
Virtual 2D-PAGE plot for 3700 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4S0E8|A0A1Y4S0E8_9FIRM Recombinase family protein OS=Flavonifractor sp. An135 OX=1965558 GN=B5E80_10555 PE=4 SV=1
MM1 pKa = 7.77TIQALNGAKK10 pKa = 8.62EE11 pKa = 4.34TVDD14 pKa = 4.6LEE16 pKa = 4.59VPYY19 pKa = 10.46DD20 pKa = 3.69PQRR23 pKa = 11.84IAILDD28 pKa = 3.88MACLDD33 pKa = 3.67ILDD36 pKa = 4.34ALGVGDD42 pKa = 4.37RR43 pKa = 11.84VVGTADD49 pKa = 2.99TSLDD53 pKa = 3.63YY54 pKa = 11.33LQDD57 pKa = 4.05YY58 pKa = 10.93INDD61 pKa = 4.76DD62 pKa = 3.54IVNLGTIKK70 pKa = 10.56EE71 pKa = 4.15ADD73 pKa = 3.82LEE75 pKa = 4.21AVMACEE81 pKa = 4.41PDD83 pKa = 3.68VIFIGGRR90 pKa = 11.84LASSYY95 pKa = 11.12DD96 pKa = 3.75ALSEE100 pKa = 4.05IAPVVYY106 pKa = 10.58LSTDD110 pKa = 3.24TEE112 pKa = 4.59LGVVEE117 pKa = 4.98SVRR120 pKa = 11.84QNATTIASMFGLEE133 pKa = 4.38DD134 pKa = 3.64KK135 pKa = 11.04VDD137 pKa = 3.67EE138 pKa = 4.88LMSDD142 pKa = 3.69FDD144 pKa = 4.24ARR146 pKa = 11.84IAALAEE152 pKa = 3.94FAAGKK157 pKa = 7.21TAIVGMCTSGSYY169 pKa = 10.48NVLGNDD175 pKa = 3.97GRR177 pKa = 11.84CSLIGVEE184 pKa = 4.98VGFEE188 pKa = 4.23NIGVDD193 pKa = 3.81ANLDD197 pKa = 3.65TSTHH201 pKa = 5.36GNEE204 pKa = 3.91ASFEE208 pKa = 4.35FIVDD212 pKa = 3.82KK213 pKa = 11.26NPDD216 pKa = 3.83YY217 pKa = 11.02IFVMDD222 pKa = 4.67RR223 pKa = 11.84DD224 pKa = 3.83AAIGTDD230 pKa = 3.96GAQLAQDD237 pKa = 3.02IMEE240 pKa = 4.74NEE242 pKa = 4.11LVMGTNAYY250 pKa = 10.27QNGHH254 pKa = 6.18LVYY257 pKa = 10.15LAHH260 pKa = 6.72PAVWYY265 pKa = 7.69TAEE268 pKa = 4.27GGITALDD275 pKa = 4.12LMLQDD280 pKa = 5.65LEE282 pKa = 4.86SEE284 pKa = 4.43LLGG287 pKa = 4.23
MM1 pKa = 7.77TIQALNGAKK10 pKa = 8.62EE11 pKa = 4.34TVDD14 pKa = 4.6LEE16 pKa = 4.59VPYY19 pKa = 10.46DD20 pKa = 3.69PQRR23 pKa = 11.84IAILDD28 pKa = 3.88MACLDD33 pKa = 3.67ILDD36 pKa = 4.34ALGVGDD42 pKa = 4.37RR43 pKa = 11.84VVGTADD49 pKa = 2.99TSLDD53 pKa = 3.63YY54 pKa = 11.33LQDD57 pKa = 4.05YY58 pKa = 10.93INDD61 pKa = 4.76DD62 pKa = 3.54IVNLGTIKK70 pKa = 10.56EE71 pKa = 4.15ADD73 pKa = 3.82LEE75 pKa = 4.21AVMACEE81 pKa = 4.41PDD83 pKa = 3.68VIFIGGRR90 pKa = 11.84LASSYY95 pKa = 11.12DD96 pKa = 3.75ALSEE100 pKa = 4.05IAPVVYY106 pKa = 10.58LSTDD110 pKa = 3.24TEE112 pKa = 4.59LGVVEE117 pKa = 4.98SVRR120 pKa = 11.84QNATTIASMFGLEE133 pKa = 4.38DD134 pKa = 3.64KK135 pKa = 11.04VDD137 pKa = 3.67EE138 pKa = 4.88LMSDD142 pKa = 3.69FDD144 pKa = 4.24ARR146 pKa = 11.84IAALAEE152 pKa = 3.94FAAGKK157 pKa = 7.21TAIVGMCTSGSYY169 pKa = 10.48NVLGNDD175 pKa = 3.97GRR177 pKa = 11.84CSLIGVEE184 pKa = 4.98VGFEE188 pKa = 4.23NIGVDD193 pKa = 3.81ANLDD197 pKa = 3.65TSTHH201 pKa = 5.36GNEE204 pKa = 3.91ASFEE208 pKa = 4.35FIVDD212 pKa = 3.82KK213 pKa = 11.26NPDD216 pKa = 3.83YY217 pKa = 11.02IFVMDD222 pKa = 4.67RR223 pKa = 11.84DD224 pKa = 3.83AAIGTDD230 pKa = 3.96GAQLAQDD237 pKa = 3.02IMEE240 pKa = 4.74NEE242 pKa = 4.11LVMGTNAYY250 pKa = 10.27QNGHH254 pKa = 6.18LVYY257 pKa = 10.15LAHH260 pKa = 6.72PAVWYY265 pKa = 7.69TAEE268 pKa = 4.27GGITALDD275 pKa = 4.12LMLQDD280 pKa = 5.65LEE282 pKa = 4.86SEE284 pKa = 4.43LLGG287 pKa = 4.23
Molecular weight: 30.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4RYG8|A0A1Y4RYG8_9FIRM Aminotransferase OS=Flavonifractor sp. An135 OX=1965558 GN=B5E80_14270 PE=3 SV=1
MM1 pKa = 7.06VRR3 pKa = 11.84TYY5 pKa = 10.22QPKK8 pKa = 9.15KK9 pKa = 7.59RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.92GFRR19 pKa = 11.84KK20 pKa = 10.08RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.05GRR39 pKa = 11.84ARR41 pKa = 11.84LSHH44 pKa = 6.55
MM1 pKa = 7.06VRR3 pKa = 11.84TYY5 pKa = 10.22QPKK8 pKa = 9.15KK9 pKa = 7.59RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.92GFRR19 pKa = 11.84KK20 pKa = 10.08RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.05GRR39 pKa = 11.84ARR41 pKa = 11.84LSHH44 pKa = 6.55
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1150233 |
25 |
2902 |
310.9 |
34.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.673 ± 0.053 | 1.628 ± 0.018 |
5.661 ± 0.033 | 7.078 ± 0.045 |
3.665 ± 0.026 | 7.959 ± 0.037 |
1.864 ± 0.019 | 5.065 ± 0.038 |
4.349 ± 0.04 | 10.404 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.583 ± 0.02 | 3.058 ± 0.027 |
4.446 ± 0.026 | 3.584 ± 0.023 |
5.95 ± 0.051 | 5.415 ± 0.03 |
5.703 ± 0.043 | 7.261 ± 0.037 |
1.206 ± 0.017 | 3.447 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
