
Sewage-associated circular DNA virus-26
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.73
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0B4UI48|A0A0B4UI48_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-26 OX=1592093 PE=3 SV=1
MM1 pKa = 7.42PKK3 pKa = 9.87KK4 pKa = 10.68GKK6 pKa = 10.56FEE8 pKa = 4.2FRR10 pKa = 11.84TKK12 pKa = 10.98AKK14 pKa = 9.97LHH16 pKa = 6.12SLTYY20 pKa = 7.63PQCDD24 pKa = 3.15IPVDD28 pKa = 4.03FMMAALKK35 pKa = 10.38KK36 pKa = 10.0IAGDD40 pKa = 3.08RR41 pKa = 11.84WGWALIGAEE50 pKa = 4.12DD51 pKa = 3.59HH52 pKa = 7.34AEE54 pKa = 3.9RR55 pKa = 11.84TTDD58 pKa = 3.39EE59 pKa = 4.45FKK61 pKa = 10.15HH62 pKa = 6.35CGVHH66 pKa = 4.41RR67 pKa = 11.84HH68 pKa = 5.52VMQQYY73 pKa = 7.93EE74 pKa = 4.84CKK76 pKa = 10.75SFDD79 pKa = 3.9TVNVRR84 pKa = 11.84YY85 pKa = 9.11WDD87 pKa = 3.68IEE89 pKa = 4.34WNGKK93 pKa = 9.02NYY95 pKa = 9.89HH96 pKa = 5.91PHH98 pKa = 6.33FEE100 pKa = 4.07PVKK103 pKa = 10.59IKK105 pKa = 10.83AKK107 pKa = 8.92WLEE110 pKa = 3.81YY111 pKa = 10.42CMKK114 pKa = 10.05DD115 pKa = 3.15GEE117 pKa = 4.56YY118 pKa = 9.75IYY120 pKa = 10.91EE121 pKa = 4.25GDD123 pKa = 3.83YY124 pKa = 11.36NGAPFDD130 pKa = 4.65PNTYY134 pKa = 9.79FQANKK139 pKa = 10.05SKK141 pKa = 10.66QSYY144 pKa = 8.72GFVWMANEE152 pKa = 3.75IKK154 pKa = 10.5AGKK157 pKa = 7.79TLDD160 pKa = 3.87EE161 pKa = 5.34LDD163 pKa = 4.42DD164 pKa = 4.2LVPGHH169 pKa = 5.9VLQHH173 pKa = 5.35KK174 pKa = 10.15RR175 pKa = 11.84KK176 pKa = 8.74IEE178 pKa = 4.06EE179 pKa = 4.01YY180 pKa = 10.86QEE182 pKa = 3.85FQAKK186 pKa = 9.55KK187 pKa = 10.08KK188 pKa = 9.09EE189 pKa = 3.88RR190 pKa = 11.84RR191 pKa = 11.84TPKK194 pKa = 9.95PIFYY198 pKa = 10.33GFNDD202 pKa = 3.47VEE204 pKa = 4.28DD205 pKa = 4.54PQWQRR210 pKa = 11.84VVKK213 pKa = 9.23WANKK217 pKa = 9.27NFLKK221 pKa = 10.59KK222 pKa = 10.27RR223 pKa = 11.84SSKK226 pKa = 10.41QKK228 pKa = 10.62QLFLWSEE235 pKa = 4.0APNLGKK241 pKa = 7.96THH243 pKa = 7.08PWAEE247 pKa = 4.04TLTEE251 pKa = 4.79FFKK254 pKa = 11.03CYY256 pKa = 9.99NWVNSEE262 pKa = 4.18RR263 pKa = 11.84QGKK266 pKa = 9.53ALMDD270 pKa = 4.13CDD272 pKa = 4.89YY273 pKa = 11.38ILLDD277 pKa = 4.28EE278 pKa = 4.62YY279 pKa = 11.43KK280 pKa = 11.13GMLTITDD287 pKa = 4.56LKK289 pKa = 10.95RR290 pKa = 11.84NSQMMKK296 pKa = 8.49WPVEE300 pKa = 3.83IKK302 pKa = 10.88YY303 pKa = 10.83GDD305 pKa = 3.52STEE308 pKa = 4.13FTRR311 pKa = 11.84NVPMIITSNHH321 pKa = 5.73DD322 pKa = 2.96MRR324 pKa = 11.84SIYY327 pKa = 10.05KK328 pKa = 9.75KK329 pKa = 10.1CDD331 pKa = 3.18YY332 pKa = 10.96RR333 pKa = 11.84DD334 pKa = 3.55IEE336 pKa = 4.29SLEE339 pKa = 3.81VRR341 pKa = 11.84FKK343 pKa = 10.69VVKK346 pKa = 10.54VDD348 pKa = 4.12TVCHH352 pKa = 5.95LTLKK356 pKa = 10.51GPPPVVLVPLPRR368 pKa = 11.84EE369 pKa = 3.99QPTLVAVPEE378 pKa = 3.98NDD380 pKa = 4.36KK381 pKa = 11.37EE382 pKa = 4.15DD383 pKa = 4.03LSDD386 pKa = 3.67EE387 pKa = 4.29EE388 pKa = 5.13EE389 pKa = 4.33ILSSLDD395 pKa = 3.97DD396 pKa = 4.18EE397 pKa = 5.68DD398 pKa = 6.07SEE400 pKa = 4.76FTTTLKK406 pKa = 10.78LAAKK410 pKa = 9.24EE411 pKa = 4.09KK412 pKa = 10.65YY413 pKa = 8.85SVKK416 pKa = 10.68YY417 pKa = 10.24FDD419 pKa = 3.85
MM1 pKa = 7.42PKK3 pKa = 9.87KK4 pKa = 10.68GKK6 pKa = 10.56FEE8 pKa = 4.2FRR10 pKa = 11.84TKK12 pKa = 10.98AKK14 pKa = 9.97LHH16 pKa = 6.12SLTYY20 pKa = 7.63PQCDD24 pKa = 3.15IPVDD28 pKa = 4.03FMMAALKK35 pKa = 10.38KK36 pKa = 10.0IAGDD40 pKa = 3.08RR41 pKa = 11.84WGWALIGAEE50 pKa = 4.12DD51 pKa = 3.59HH52 pKa = 7.34AEE54 pKa = 3.9RR55 pKa = 11.84TTDD58 pKa = 3.39EE59 pKa = 4.45FKK61 pKa = 10.15HH62 pKa = 6.35CGVHH66 pKa = 4.41RR67 pKa = 11.84HH68 pKa = 5.52VMQQYY73 pKa = 7.93EE74 pKa = 4.84CKK76 pKa = 10.75SFDD79 pKa = 3.9TVNVRR84 pKa = 11.84YY85 pKa = 9.11WDD87 pKa = 3.68IEE89 pKa = 4.34WNGKK93 pKa = 9.02NYY95 pKa = 9.89HH96 pKa = 5.91PHH98 pKa = 6.33FEE100 pKa = 4.07PVKK103 pKa = 10.59IKK105 pKa = 10.83AKK107 pKa = 8.92WLEE110 pKa = 3.81YY111 pKa = 10.42CMKK114 pKa = 10.05DD115 pKa = 3.15GEE117 pKa = 4.56YY118 pKa = 9.75IYY120 pKa = 10.91EE121 pKa = 4.25GDD123 pKa = 3.83YY124 pKa = 11.36NGAPFDD130 pKa = 4.65PNTYY134 pKa = 9.79FQANKK139 pKa = 10.05SKK141 pKa = 10.66QSYY144 pKa = 8.72GFVWMANEE152 pKa = 3.75IKK154 pKa = 10.5AGKK157 pKa = 7.79TLDD160 pKa = 3.87EE161 pKa = 5.34LDD163 pKa = 4.42DD164 pKa = 4.2LVPGHH169 pKa = 5.9VLQHH173 pKa = 5.35KK174 pKa = 10.15RR175 pKa = 11.84KK176 pKa = 8.74IEE178 pKa = 4.06EE179 pKa = 4.01YY180 pKa = 10.86QEE182 pKa = 3.85FQAKK186 pKa = 9.55KK187 pKa = 10.08KK188 pKa = 9.09EE189 pKa = 3.88RR190 pKa = 11.84RR191 pKa = 11.84TPKK194 pKa = 9.95PIFYY198 pKa = 10.33GFNDD202 pKa = 3.47VEE204 pKa = 4.28DD205 pKa = 4.54PQWQRR210 pKa = 11.84VVKK213 pKa = 9.23WANKK217 pKa = 9.27NFLKK221 pKa = 10.59KK222 pKa = 10.27RR223 pKa = 11.84SSKK226 pKa = 10.41QKK228 pKa = 10.62QLFLWSEE235 pKa = 4.0APNLGKK241 pKa = 7.96THH243 pKa = 7.08PWAEE247 pKa = 4.04TLTEE251 pKa = 4.79FFKK254 pKa = 11.03CYY256 pKa = 9.99NWVNSEE262 pKa = 4.18RR263 pKa = 11.84QGKK266 pKa = 9.53ALMDD270 pKa = 4.13CDD272 pKa = 4.89YY273 pKa = 11.38ILLDD277 pKa = 4.28EE278 pKa = 4.62YY279 pKa = 11.43KK280 pKa = 11.13GMLTITDD287 pKa = 4.56LKK289 pKa = 10.95RR290 pKa = 11.84NSQMMKK296 pKa = 8.49WPVEE300 pKa = 3.83IKK302 pKa = 10.88YY303 pKa = 10.83GDD305 pKa = 3.52STEE308 pKa = 4.13FTRR311 pKa = 11.84NVPMIITSNHH321 pKa = 5.73DD322 pKa = 2.96MRR324 pKa = 11.84SIYY327 pKa = 10.05KK328 pKa = 9.75KK329 pKa = 10.1CDD331 pKa = 3.18YY332 pKa = 10.96RR333 pKa = 11.84DD334 pKa = 3.55IEE336 pKa = 4.29SLEE339 pKa = 3.81VRR341 pKa = 11.84FKK343 pKa = 10.69VVKK346 pKa = 10.54VDD348 pKa = 4.12TVCHH352 pKa = 5.95LTLKK356 pKa = 10.51GPPPVVLVPLPRR368 pKa = 11.84EE369 pKa = 3.99QPTLVAVPEE378 pKa = 3.98NDD380 pKa = 4.36KK381 pKa = 11.37EE382 pKa = 4.15DD383 pKa = 4.03LSDD386 pKa = 3.67EE387 pKa = 4.29EE388 pKa = 5.13EE389 pKa = 4.33ILSSLDD395 pKa = 3.97DD396 pKa = 4.18EE397 pKa = 5.68DD398 pKa = 6.07SEE400 pKa = 4.76FTTTLKK406 pKa = 10.78LAAKK410 pKa = 9.24EE411 pKa = 4.09KK412 pKa = 10.65YY413 pKa = 8.85SVKK416 pKa = 10.68YY417 pKa = 10.24FDD419 pKa = 3.85
Molecular weight: 49.41 kDa
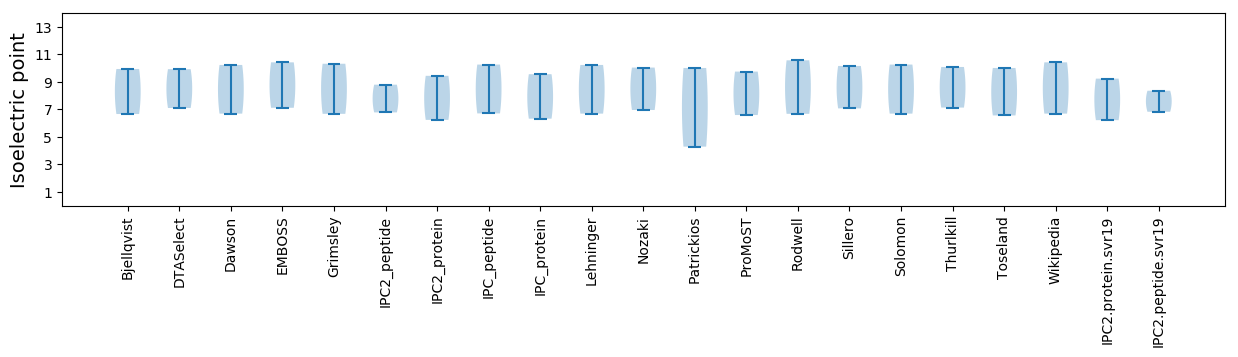
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI48|A0A0B4UI48_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-26 OX=1592093 PE=3 SV=1
MM1 pKa = 7.58SYY3 pKa = 10.78KK4 pKa = 10.6SKK6 pKa = 10.57LWVGKK11 pKa = 10.19RR12 pKa = 11.84NAPRR16 pKa = 11.84TKK18 pKa = 9.33SQRR21 pKa = 11.84YY22 pKa = 7.86ANQVRR27 pKa = 11.84TNLLRR32 pKa = 11.84KK33 pKa = 9.34IVGNRR38 pKa = 11.84VPRR41 pKa = 11.84PMPMRR46 pKa = 11.84RR47 pKa = 11.84SIKK50 pKa = 9.6TGEE53 pKa = 4.27IKK55 pKa = 10.89CLDD58 pKa = 3.49QTFLATYY65 pKa = 9.14NATWVPDD72 pKa = 3.8AFPQKK77 pKa = 10.65VLNADD82 pKa = 3.54SNGAFQAINLIQQGNGISQRR102 pKa = 11.84IGNKK106 pKa = 9.41INLKK110 pKa = 8.4SVRR113 pKa = 11.84LRR115 pKa = 11.84FSLDD119 pKa = 3.0PTTVTTASPTDD130 pKa = 3.44TRR132 pKa = 11.84VMLVYY137 pKa = 10.47DD138 pKa = 4.2RR139 pKa = 11.84QPNGLYY145 pKa = 10.1PSLNDD150 pKa = 3.33FLNGVSQSNALLNPSGLALCSSNINPNNFEE180 pKa = 4.21RR181 pKa = 11.84FTVLMDD187 pKa = 3.88KK188 pKa = 10.87SITLPSSQASSNFGPTGDD206 pKa = 4.37GQNSPWWIDD215 pKa = 3.46EE216 pKa = 4.28YY217 pKa = 10.82IKK219 pKa = 11.1CKK221 pKa = 10.24GLPVTYY227 pKa = 9.64KK228 pKa = 10.75ANSAPSVIGDD238 pKa = 3.63IATGTLFLFCVGEE251 pKa = 4.09NVALGTSWSYY261 pKa = 10.97RR262 pKa = 11.84GQARR266 pKa = 11.84LRR268 pKa = 11.84YY269 pKa = 9.11YY270 pKa = 10.77DD271 pKa = 3.22AA272 pKa = 6.64
MM1 pKa = 7.58SYY3 pKa = 10.78KK4 pKa = 10.6SKK6 pKa = 10.57LWVGKK11 pKa = 10.19RR12 pKa = 11.84NAPRR16 pKa = 11.84TKK18 pKa = 9.33SQRR21 pKa = 11.84YY22 pKa = 7.86ANQVRR27 pKa = 11.84TNLLRR32 pKa = 11.84KK33 pKa = 9.34IVGNRR38 pKa = 11.84VPRR41 pKa = 11.84PMPMRR46 pKa = 11.84RR47 pKa = 11.84SIKK50 pKa = 9.6TGEE53 pKa = 4.27IKK55 pKa = 10.89CLDD58 pKa = 3.49QTFLATYY65 pKa = 9.14NATWVPDD72 pKa = 3.8AFPQKK77 pKa = 10.65VLNADD82 pKa = 3.54SNGAFQAINLIQQGNGISQRR102 pKa = 11.84IGNKK106 pKa = 9.41INLKK110 pKa = 8.4SVRR113 pKa = 11.84LRR115 pKa = 11.84FSLDD119 pKa = 3.0PTTVTTASPTDD130 pKa = 3.44TRR132 pKa = 11.84VMLVYY137 pKa = 10.47DD138 pKa = 4.2RR139 pKa = 11.84QPNGLYY145 pKa = 10.1PSLNDD150 pKa = 3.33FLNGVSQSNALLNPSGLALCSSNINPNNFEE180 pKa = 4.21RR181 pKa = 11.84FTVLMDD187 pKa = 3.88KK188 pKa = 10.87SITLPSSQASSNFGPTGDD206 pKa = 4.37GQNSPWWIDD215 pKa = 3.46EE216 pKa = 4.28YY217 pKa = 10.82IKK219 pKa = 11.1CKK221 pKa = 10.24GLPVTYY227 pKa = 9.64KK228 pKa = 10.75ANSAPSVIGDD238 pKa = 3.63IATGTLFLFCVGEE251 pKa = 4.09NVALGTSWSYY261 pKa = 10.97RR262 pKa = 11.84GQARR266 pKa = 11.84LRR268 pKa = 11.84YY269 pKa = 9.11YY270 pKa = 10.77DD271 pKa = 3.22AA272 pKa = 6.64
Molecular weight: 30.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
691 |
272 |
419 |
345.5 |
39.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.499 ± 0.638 | 1.737 ± 0.152 |
6.223 ± 1.033 | 5.644 ± 2.381 |
4.342 ± 0.379 | 5.355 ± 0.93 |
1.737 ± 0.991 | 4.486 ± 0.377 |
8.828 ± 2.1 | 7.959 ± 0.703 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.46 ± 0.355 | 5.933 ± 1.649 |
5.644 ± 0.346 | 4.052 ± 0.415 |
5.065 ± 0.886 | 6.223 ± 1.694 |
5.933 ± 0.6 | 6.078 ± 0.112 |
2.46 ± 0.355 | 4.342 ± 0.379 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
