
Chimpanzee associated porprismacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
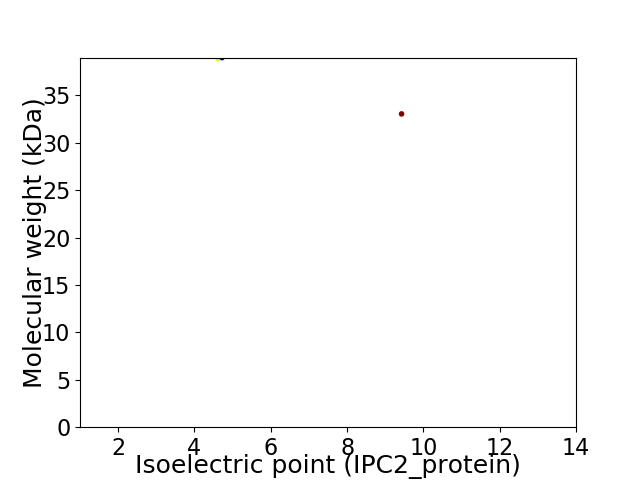
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2WKE3|D2WKE3_9VIRU Putative replicase protein OS=Chimpanzee associated porprismacovirus 1 OX=2170109 GN=rep PE=4 SV=1
MM1 pKa = 7.01FVKK4 pKa = 10.65VSEE7 pKa = 4.49TYY9 pKa = 10.72DD10 pKa = 4.37LSTQTDD16 pKa = 2.71KK17 pKa = 11.16MGFVGIHH24 pKa = 4.75TPEE27 pKa = 4.3GKK29 pKa = 9.88LVYY32 pKa = 10.76NMWSGLFKK40 pKa = 10.78NFRR43 pKa = 11.84KK44 pKa = 9.39FRR46 pKa = 11.84YY47 pKa = 8.92ASCDD51 pKa = 3.08VTMACASMLPADD63 pKa = 4.26PLQIGVEE70 pKa = 4.09AGDD73 pKa = 4.21IAPQDD78 pKa = 3.63MFNPILYY85 pKa = 8.86KK86 pKa = 10.64ACSNDD91 pKa = 3.26SMSVLLNRR99 pKa = 11.84LYY101 pKa = 11.07AGADD105 pKa = 3.19ASTSAINKK113 pKa = 9.21NSVVAQNDD121 pKa = 3.17EE122 pKa = 4.44SFGYY126 pKa = 10.62DD127 pKa = 3.07ADD129 pKa = 4.02NDD131 pKa = 3.39VDD133 pKa = 4.06QFAMYY138 pKa = 10.28YY139 pKa = 10.82GLLADD144 pKa = 3.49SDD146 pKa = 4.19GWRR149 pKa = 11.84KK150 pKa = 10.0AMPQAGLQMHH160 pKa = 6.13GRR162 pKa = 11.84KK163 pKa = 8.97PLVFGITSNFGQPSNVGLPNGKK185 pKa = 8.15MVYY188 pKa = 9.01TGNGDD193 pKa = 3.34GSLTARR199 pKa = 11.84NTIDD203 pKa = 3.26STLFQYY209 pKa = 9.91MRR211 pKa = 11.84GPTMGMPALDD221 pKa = 3.54TFVQLGGSTVVNGFADD237 pKa = 3.74SVTVPEE243 pKa = 4.4VSDD246 pKa = 3.29RR247 pKa = 11.84AYY249 pKa = 9.87PISNGSITTPGNVCTPNLDD268 pKa = 3.78TPDD271 pKa = 3.94CFVAAIVLPPAKK283 pKa = 10.22LNRR286 pKa = 11.84LYY288 pKa = 11.4YY289 pKa = 10.02RR290 pKa = 11.84MKK292 pKa = 9.55VTWTIEE298 pKa = 4.11FSGLRR303 pKa = 11.84PDD305 pKa = 3.7TDD307 pKa = 3.35LTNWYY312 pKa = 9.43GLALAGVQSYY322 pKa = 10.98GSDD325 pKa = 3.47YY326 pKa = 10.38LTQTATIARR335 pKa = 11.84ATSTANTSGMVDD347 pKa = 3.1TGDD350 pKa = 3.35VSLTKK355 pKa = 10.74VMEE358 pKa = 4.68GASS361 pKa = 3.42
MM1 pKa = 7.01FVKK4 pKa = 10.65VSEE7 pKa = 4.49TYY9 pKa = 10.72DD10 pKa = 4.37LSTQTDD16 pKa = 2.71KK17 pKa = 11.16MGFVGIHH24 pKa = 4.75TPEE27 pKa = 4.3GKK29 pKa = 9.88LVYY32 pKa = 10.76NMWSGLFKK40 pKa = 10.78NFRR43 pKa = 11.84KK44 pKa = 9.39FRR46 pKa = 11.84YY47 pKa = 8.92ASCDD51 pKa = 3.08VTMACASMLPADD63 pKa = 4.26PLQIGVEE70 pKa = 4.09AGDD73 pKa = 4.21IAPQDD78 pKa = 3.63MFNPILYY85 pKa = 8.86KK86 pKa = 10.64ACSNDD91 pKa = 3.26SMSVLLNRR99 pKa = 11.84LYY101 pKa = 11.07AGADD105 pKa = 3.19ASTSAINKK113 pKa = 9.21NSVVAQNDD121 pKa = 3.17EE122 pKa = 4.44SFGYY126 pKa = 10.62DD127 pKa = 3.07ADD129 pKa = 4.02NDD131 pKa = 3.39VDD133 pKa = 4.06QFAMYY138 pKa = 10.28YY139 pKa = 10.82GLLADD144 pKa = 3.49SDD146 pKa = 4.19GWRR149 pKa = 11.84KK150 pKa = 10.0AMPQAGLQMHH160 pKa = 6.13GRR162 pKa = 11.84KK163 pKa = 8.97PLVFGITSNFGQPSNVGLPNGKK185 pKa = 8.15MVYY188 pKa = 9.01TGNGDD193 pKa = 3.34GSLTARR199 pKa = 11.84NTIDD203 pKa = 3.26STLFQYY209 pKa = 9.91MRR211 pKa = 11.84GPTMGMPALDD221 pKa = 3.54TFVQLGGSTVVNGFADD237 pKa = 3.74SVTVPEE243 pKa = 4.4VSDD246 pKa = 3.29RR247 pKa = 11.84AYY249 pKa = 9.87PISNGSITTPGNVCTPNLDD268 pKa = 3.78TPDD271 pKa = 3.94CFVAAIVLPPAKK283 pKa = 10.22LNRR286 pKa = 11.84LYY288 pKa = 11.4YY289 pKa = 10.02RR290 pKa = 11.84MKK292 pKa = 9.55VTWTIEE298 pKa = 4.11FSGLRR303 pKa = 11.84PDD305 pKa = 3.7TDD307 pKa = 3.35LTNWYY312 pKa = 9.43GLALAGVQSYY322 pKa = 10.98GSDD325 pKa = 3.47YY326 pKa = 10.38LTQTATIARR335 pKa = 11.84ATSTANTSGMVDD347 pKa = 3.1TGDD350 pKa = 3.35VSLTKK355 pKa = 10.74VMEE358 pKa = 4.68GASS361 pKa = 3.42
Molecular weight: 38.88 kDa
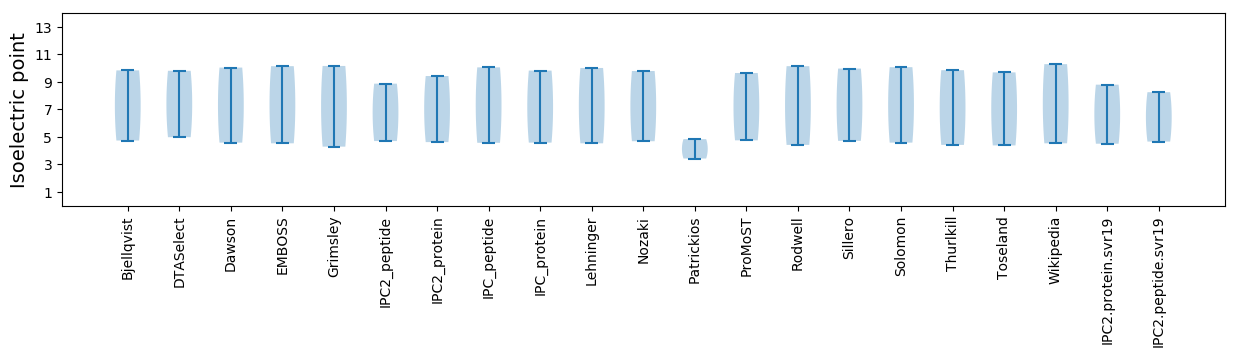
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2WKE3|D2WKE3_9VIRU Putative replicase protein OS=Chimpanzee associated porprismacovirus 1 OX=2170109 GN=rep PE=4 SV=1
MM1 pKa = 7.3GHH3 pKa = 5.99MFGYY7 pKa = 8.5TCLKK11 pKa = 9.54RR12 pKa = 11.84GCPRR16 pKa = 11.84RR17 pKa = 11.84PLDD20 pKa = 3.25YY21 pKa = 11.08ALVYY25 pKa = 10.52GVGISGHH32 pKa = 5.9NDD34 pKa = 2.9NDD36 pKa = 3.65GTHH39 pKa = 5.19PHH41 pKa = 6.02EE42 pKa = 4.72RR43 pKa = 11.84RR44 pKa = 11.84ANPLVQNLPRR54 pKa = 11.84SRR56 pKa = 11.84YY57 pKa = 8.26SQMGYY62 pKa = 9.79RR63 pKa = 11.84AGGRR67 pKa = 11.84SRR69 pKa = 11.84RR70 pKa = 11.84VQALAGQMQCAGCEE84 pKa = 3.8RR85 pKa = 11.84PAVDD89 pKa = 3.38GLSTVRR95 pKa = 11.84IRR97 pKa = 11.84MGRR100 pKa = 11.84GYY102 pKa = 10.44LDD104 pKa = 3.16SYY106 pKa = 10.03RR107 pKa = 11.84RR108 pKa = 11.84SVRR111 pKa = 11.84ISTSTRR117 pKa = 11.84RR118 pKa = 11.84KK119 pKa = 9.34RR120 pKa = 11.84ATIGHH125 pKa = 6.55RR126 pKa = 11.84GIRR129 pKa = 11.84WAHH132 pKa = 5.33VNSEE136 pKa = 4.13FGKK139 pKa = 9.44MRR141 pKa = 11.84WNQEE145 pKa = 3.43GALEE149 pKa = 4.1ALQRR153 pKa = 11.84TNDD156 pKa = 3.41RR157 pKa = 11.84EE158 pKa = 4.08IVVWYY163 pKa = 9.81DD164 pKa = 2.92QDD166 pKa = 4.05GNMGKK171 pKa = 9.23SWLCGHH177 pKa = 7.6LYY179 pKa = 8.32EE180 pKa = 5.0TGKK183 pKa = 10.73AYY185 pKa = 10.16YY186 pKa = 8.97IPPYY190 pKa = 10.09ISTIQSMIQTVASLVLQDD208 pKa = 4.91RR209 pKa = 11.84EE210 pKa = 3.99AGYY213 pKa = 9.36PPRR216 pKa = 11.84PLIVIDD222 pKa = 5.23IPRR225 pKa = 11.84SWKK228 pKa = 9.63WSTEE232 pKa = 3.64LYY234 pKa = 9.64TAIEE238 pKa = 4.54AIKK241 pKa = 10.41DD242 pKa = 3.74GLIMDD247 pKa = 4.67PRR249 pKa = 11.84YY250 pKa = 9.71GARR253 pKa = 11.84PVNIHH258 pKa = 4.49GTKK261 pKa = 10.52VIVLTNTKK269 pKa = 9.98PKK271 pKa = 10.2LDD273 pKa = 3.97KK274 pKa = 10.88LSEE277 pKa = 4.3DD278 pKa = 2.82RR279 pKa = 11.84WVLYY283 pKa = 10.74DD284 pKa = 3.75PLLYY288 pKa = 9.93MM289 pKa = 5.73
MM1 pKa = 7.3GHH3 pKa = 5.99MFGYY7 pKa = 8.5TCLKK11 pKa = 9.54RR12 pKa = 11.84GCPRR16 pKa = 11.84RR17 pKa = 11.84PLDD20 pKa = 3.25YY21 pKa = 11.08ALVYY25 pKa = 10.52GVGISGHH32 pKa = 5.9NDD34 pKa = 2.9NDD36 pKa = 3.65GTHH39 pKa = 5.19PHH41 pKa = 6.02EE42 pKa = 4.72RR43 pKa = 11.84RR44 pKa = 11.84ANPLVQNLPRR54 pKa = 11.84SRR56 pKa = 11.84YY57 pKa = 8.26SQMGYY62 pKa = 9.79RR63 pKa = 11.84AGGRR67 pKa = 11.84SRR69 pKa = 11.84RR70 pKa = 11.84VQALAGQMQCAGCEE84 pKa = 3.8RR85 pKa = 11.84PAVDD89 pKa = 3.38GLSTVRR95 pKa = 11.84IRR97 pKa = 11.84MGRR100 pKa = 11.84GYY102 pKa = 10.44LDD104 pKa = 3.16SYY106 pKa = 10.03RR107 pKa = 11.84RR108 pKa = 11.84SVRR111 pKa = 11.84ISTSTRR117 pKa = 11.84RR118 pKa = 11.84KK119 pKa = 9.34RR120 pKa = 11.84ATIGHH125 pKa = 6.55RR126 pKa = 11.84GIRR129 pKa = 11.84WAHH132 pKa = 5.33VNSEE136 pKa = 4.13FGKK139 pKa = 9.44MRR141 pKa = 11.84WNQEE145 pKa = 3.43GALEE149 pKa = 4.1ALQRR153 pKa = 11.84TNDD156 pKa = 3.41RR157 pKa = 11.84EE158 pKa = 4.08IVVWYY163 pKa = 9.81DD164 pKa = 2.92QDD166 pKa = 4.05GNMGKK171 pKa = 9.23SWLCGHH177 pKa = 7.6LYY179 pKa = 8.32EE180 pKa = 5.0TGKK183 pKa = 10.73AYY185 pKa = 10.16YY186 pKa = 8.97IPPYY190 pKa = 10.09ISTIQSMIQTVASLVLQDD208 pKa = 4.91RR209 pKa = 11.84EE210 pKa = 3.99AGYY213 pKa = 9.36PPRR216 pKa = 11.84PLIVIDD222 pKa = 5.23IPRR225 pKa = 11.84SWKK228 pKa = 9.63WSTEE232 pKa = 3.64LYY234 pKa = 9.64TAIEE238 pKa = 4.54AIKK241 pKa = 10.41DD242 pKa = 3.74GLIMDD247 pKa = 4.67PRR249 pKa = 11.84YY250 pKa = 9.71GARR253 pKa = 11.84PVNIHH258 pKa = 4.49GTKK261 pKa = 10.52VIVLTNTKK269 pKa = 9.98PKK271 pKa = 10.2LDD273 pKa = 3.97KK274 pKa = 10.88LSEE277 pKa = 4.3DD278 pKa = 2.82RR279 pKa = 11.84WVLYY283 pKa = 10.74DD284 pKa = 3.75PLLYY288 pKa = 9.93MM289 pKa = 5.73
Molecular weight: 33.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
650 |
289 |
361 |
325.0 |
35.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.385 ± 0.99 | 1.538 ± 0.126 |
6.462 ± 0.838 | 2.769 ± 0.683 |
2.615 ± 1.267 | 9.231 ± 0.074 |
1.538 ± 0.81 | 4.615 ± 1.063 |
3.692 ± 0.075 | 7.846 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.154 ± 0.457 | 4.769 ± 0.863 |
5.385 ± 0.1 | 3.538 ± 0.176 |
6.769 ± 2.836 | 7.077 ± 0.787 |
7.077 ± 1.243 | 6.769 ± 0.584 |
1.692 ± 0.481 | 5.077 ± 0.531 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
