
Nootka lupine vein clearing virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Alphacarmovirus
Average proteome isoelectric point is 8.54
Get precalculated fractions of proteins
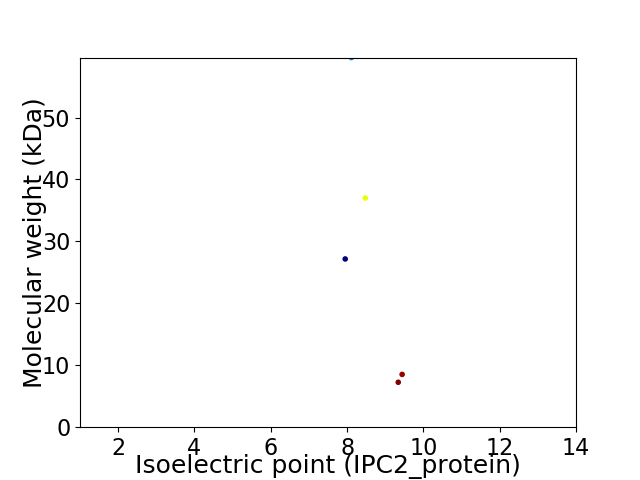
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A2TJS9|A2TJS9_9TOMB RNA-directed RNA polymerase (Fragment) OS=Nootka lupine vein clearing virus OX=283876 PE=4 SV=1
MM1 pKa = 7.49GVFSKK6 pKa = 10.56LAKK9 pKa = 9.92EE10 pKa = 3.92LVIGSGTLGVLGCCLYY26 pKa = 10.46AAAVVRR32 pKa = 11.84VTVGTVEE39 pKa = 4.11FGIAAVNTVSDD50 pKa = 4.72FVQSGGLSIPTVPQYY65 pKa = 11.36PNLGYY70 pKa = 10.71SPFQVEE76 pKa = 4.83LDD78 pKa = 3.54QSAGKK83 pKa = 9.5EE84 pKa = 3.95DD85 pKa = 3.95EE86 pKa = 5.08GISMLEE92 pKa = 3.67ASTLEE97 pKa = 4.2EE98 pKa = 4.09KK99 pKa = 10.85KK100 pKa = 10.56EE101 pKa = 3.79ADD103 pKa = 3.54SEE105 pKa = 4.54GKK107 pKa = 9.07MVVVSRR113 pKa = 11.84KK114 pKa = 9.36RR115 pKa = 11.84KK116 pKa = 8.72VNRR119 pKa = 11.84HH120 pKa = 4.35QKK122 pKa = 9.16GQFVMDD128 pKa = 3.76VVRR131 pKa = 11.84KK132 pKa = 9.8AKK134 pKa = 9.28MHH136 pKa = 6.37FGCTPSPTRR145 pKa = 11.84ANEE148 pKa = 3.83LAVAKK153 pKa = 10.04YY154 pKa = 10.21VAGLCKK160 pKa = 9.81EE161 pKa = 3.92QHH163 pKa = 6.12LVASHH168 pKa = 5.71ARR170 pKa = 11.84EE171 pKa = 3.92VTEE174 pKa = 3.93VAKK177 pKa = 10.97ALVFTPDD184 pKa = 2.77IAEE187 pKa = 4.01IQGTKK192 pKa = 10.46LLNSYY197 pKa = 9.55RR198 pKa = 11.84AYY200 pKa = 10.09ARR202 pKa = 11.84RR203 pKa = 11.84VALHH207 pKa = 7.1DD208 pKa = 4.19AQQIDD213 pKa = 4.27QWWLNLVKK221 pKa = 10.65HH222 pKa = 5.99PLSTNSWAEE231 pKa = 3.11AWYY234 pKa = 8.41TLNGAPPRR242 pKa = 11.84AAVQFNKK249 pKa = 10.66
MM1 pKa = 7.49GVFSKK6 pKa = 10.56LAKK9 pKa = 9.92EE10 pKa = 3.92LVIGSGTLGVLGCCLYY26 pKa = 10.46AAAVVRR32 pKa = 11.84VTVGTVEE39 pKa = 4.11FGIAAVNTVSDD50 pKa = 4.72FVQSGGLSIPTVPQYY65 pKa = 11.36PNLGYY70 pKa = 10.71SPFQVEE76 pKa = 4.83LDD78 pKa = 3.54QSAGKK83 pKa = 9.5EE84 pKa = 3.95DD85 pKa = 3.95EE86 pKa = 5.08GISMLEE92 pKa = 3.67ASTLEE97 pKa = 4.2EE98 pKa = 4.09KK99 pKa = 10.85KK100 pKa = 10.56EE101 pKa = 3.79ADD103 pKa = 3.54SEE105 pKa = 4.54GKK107 pKa = 9.07MVVVSRR113 pKa = 11.84KK114 pKa = 9.36RR115 pKa = 11.84KK116 pKa = 8.72VNRR119 pKa = 11.84HH120 pKa = 4.35QKK122 pKa = 9.16GQFVMDD128 pKa = 3.76VVRR131 pKa = 11.84KK132 pKa = 9.8AKK134 pKa = 9.28MHH136 pKa = 6.37FGCTPSPTRR145 pKa = 11.84ANEE148 pKa = 3.83LAVAKK153 pKa = 10.04YY154 pKa = 10.21VAGLCKK160 pKa = 9.81EE161 pKa = 3.92QHH163 pKa = 6.12LVASHH168 pKa = 5.71ARR170 pKa = 11.84EE171 pKa = 3.92VTEE174 pKa = 3.93VAKK177 pKa = 10.97ALVFTPDD184 pKa = 2.77IAEE187 pKa = 4.01IQGTKK192 pKa = 10.46LLNSYY197 pKa = 9.55RR198 pKa = 11.84AYY200 pKa = 10.09ARR202 pKa = 11.84RR203 pKa = 11.84VALHH207 pKa = 7.1DD208 pKa = 4.19AQQIDD213 pKa = 4.27QWWLNLVKK221 pKa = 10.65HH222 pKa = 5.99PLSTNSWAEE231 pKa = 3.11AWYY234 pKa = 8.41TLNGAPPRR242 pKa = 11.84AAVQFNKK249 pKa = 10.66
Molecular weight: 27.16 kDa
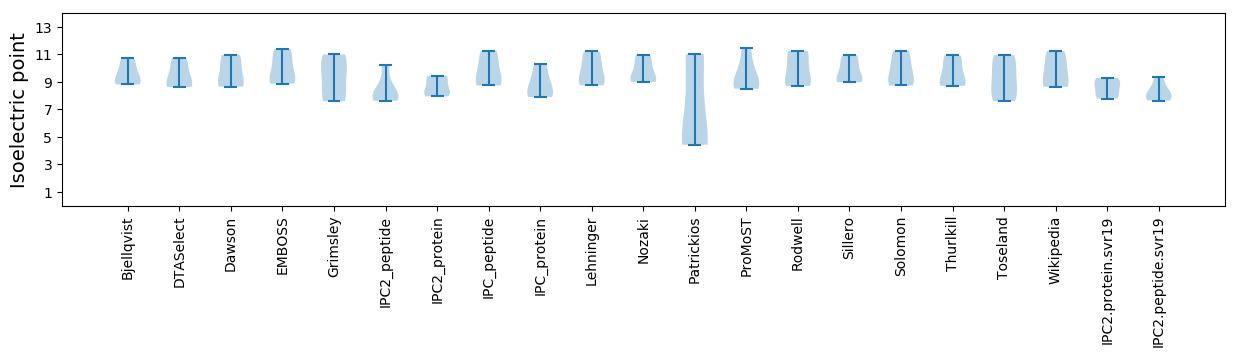
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A2TJT2|A2TJT2_9TOMB Capsid protein OS=Nootka lupine vein clearing virus OX=283876 PE=3 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84LWIKK6 pKa = 8.87MHH8 pKa = 6.15LRR10 pKa = 11.84VIVGVVTFLLLTRR23 pKa = 11.84WKK25 pKa = 9.99FAIRR29 pKa = 11.84SFSISEE35 pKa = 4.0ILPPLITLNQIITLSFLILFVNCLCRR61 pKa = 11.84GGDD64 pKa = 3.91TIFLDD69 pKa = 3.88PLHH72 pKa = 6.71RR73 pKa = 5.4
MM1 pKa = 7.5RR2 pKa = 11.84LWIKK6 pKa = 8.87MHH8 pKa = 6.15LRR10 pKa = 11.84VIVGVVTFLLLTRR23 pKa = 11.84WKK25 pKa = 9.99FAIRR29 pKa = 11.84SFSISEE35 pKa = 4.0ILPPLITLNQIITLSFLILFVNCLCRR61 pKa = 11.84GGDD64 pKa = 3.91TIFLDD69 pKa = 3.88PLHH72 pKa = 6.71RR73 pKa = 5.4
Molecular weight: 8.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1259 |
67 |
525 |
251.8 |
27.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.546 ± 1.444 | 1.986 ± 0.678 |
4.289 ± 0.444 | 5.481 ± 1.078 |
4.289 ± 0.706 | 8.181 ± 0.747 |
2.224 ± 0.284 | 4.686 ± 1.564 |
5.957 ± 1.143 | 7.466 ± 2.157 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.78 ± 0.253 | 4.21 ± 0.342 |
4.051 ± 0.129 | 3.892 ± 0.64 |
6.593 ± 0.739 | 7.069 ± 1.503 |
5.878 ± 1.234 | 9.373 ± 1.013 |
1.35 ± 0.251 | 2.701 ± 0.674 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
