
Persimmon virus A
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Persimmon cytorhabdovirus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
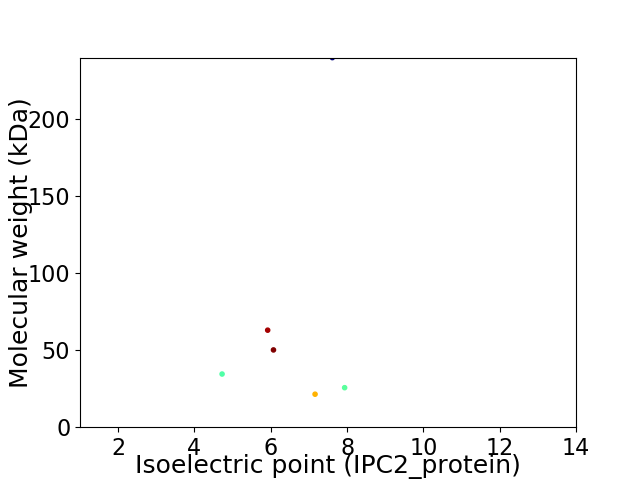
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R4WAK7|R4WAK7_9RHAB Movement protein BC1 OS=Persimmon virus A OX=1211480 PE=3 SV=1
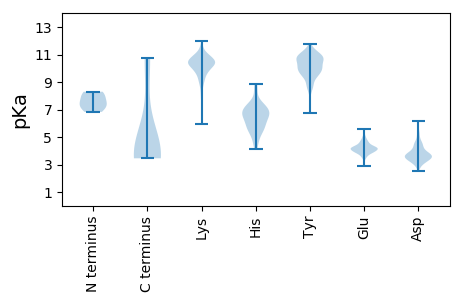
MM1 pKa = 7.7GDD3 pKa = 3.55SDD5 pKa = 3.89KK6 pKa = 11.48VNYY9 pKa = 10.17SGIGNPVGDD18 pKa = 4.05YY19 pKa = 10.78DD20 pKa = 4.56YY21 pKa = 11.5VGHH24 pKa = 6.93NSGDD28 pKa = 3.41DD29 pKa = 3.71SYY31 pKa = 11.01TGEE34 pKa = 4.16HH35 pKa = 5.17VVEE38 pKa = 4.48SVSVPEE44 pKa = 3.97VHH46 pKa = 6.68NEE48 pKa = 3.35NAVADD53 pKa = 3.68KK54 pKa = 10.43RR55 pKa = 11.84ARR57 pKa = 11.84VTPAKK62 pKa = 10.48DD63 pKa = 3.46VISWLSLHH71 pKa = 6.09SKK73 pKa = 10.42RR74 pKa = 11.84GGLVIPVEE82 pKa = 4.22MKK84 pKa = 10.69NYY86 pKa = 10.07LLSLSAEE93 pKa = 4.15DD94 pKa = 4.47LMEE97 pKa = 4.35EE98 pKa = 4.2RR99 pKa = 11.84DD100 pKa = 3.31VEE102 pKa = 4.47MFVKK106 pKa = 10.6GYY108 pKa = 8.04VFCSSSRR115 pKa = 11.84IIPNMQSVVNDD126 pKa = 3.27IQAEE130 pKa = 4.22VRR132 pKa = 11.84NLQRR136 pKa = 11.84EE137 pKa = 4.34SNKK140 pKa = 10.13NVDD143 pKa = 3.46ILRR146 pKa = 11.84SMEE149 pKa = 4.01KK150 pKa = 9.8QGKK153 pKa = 5.94STEE156 pKa = 4.05MEE158 pKa = 3.51IAAAVATIRR167 pKa = 11.84SDD169 pKa = 2.9MVTAIKK175 pKa = 10.4SALKK179 pKa = 9.87EE180 pKa = 3.95QVTATSSSSGKK191 pKa = 9.92VEE193 pKa = 3.87LKK195 pKa = 10.81VRR197 pKa = 11.84TEE199 pKa = 3.86KK200 pKa = 10.71QAGLDD205 pKa = 3.69VKK207 pKa = 10.17QVPLLINPTTSKK219 pKa = 10.86LPRR222 pKa = 11.84KK223 pKa = 9.09PDD225 pKa = 3.45LEE227 pKa = 4.07AVRR230 pKa = 11.84PSSSKK235 pKa = 10.12SSKK238 pKa = 10.24EE239 pKa = 3.68VTLNEE244 pKa = 3.97LKK246 pKa = 10.54RR247 pKa = 11.84IFMCAVGVEE256 pKa = 3.86ADD258 pKa = 3.44IVSVLTDD265 pKa = 3.57EE266 pKa = 5.35EE267 pKa = 4.36VDD269 pKa = 4.66AMLTANDD276 pKa = 4.01YY277 pKa = 11.76YY278 pKa = 10.67EE279 pKa = 4.3FCNDD283 pKa = 3.76LDD285 pKa = 4.53DD286 pKa = 5.81DD287 pKa = 4.49GVRR290 pKa = 11.84KK291 pKa = 9.64EE292 pKa = 4.04FSEE295 pKa = 4.16IFNKK299 pKa = 10.91AIDD302 pKa = 3.93QLNKK306 pKa = 9.94EE307 pKa = 4.51AEE309 pKa = 4.26EE310 pKa = 4.1EE311 pKa = 4.22DD312 pKa = 3.65
MM1 pKa = 7.7GDD3 pKa = 3.55SDD5 pKa = 3.89KK6 pKa = 11.48VNYY9 pKa = 10.17SGIGNPVGDD18 pKa = 4.05YY19 pKa = 10.78DD20 pKa = 4.56YY21 pKa = 11.5VGHH24 pKa = 6.93NSGDD28 pKa = 3.41DD29 pKa = 3.71SYY31 pKa = 11.01TGEE34 pKa = 4.16HH35 pKa = 5.17VVEE38 pKa = 4.48SVSVPEE44 pKa = 3.97VHH46 pKa = 6.68NEE48 pKa = 3.35NAVADD53 pKa = 3.68KK54 pKa = 10.43RR55 pKa = 11.84ARR57 pKa = 11.84VTPAKK62 pKa = 10.48DD63 pKa = 3.46VISWLSLHH71 pKa = 6.09SKK73 pKa = 10.42RR74 pKa = 11.84GGLVIPVEE82 pKa = 4.22MKK84 pKa = 10.69NYY86 pKa = 10.07LLSLSAEE93 pKa = 4.15DD94 pKa = 4.47LMEE97 pKa = 4.35EE98 pKa = 4.2RR99 pKa = 11.84DD100 pKa = 3.31VEE102 pKa = 4.47MFVKK106 pKa = 10.6GYY108 pKa = 8.04VFCSSSRR115 pKa = 11.84IIPNMQSVVNDD126 pKa = 3.27IQAEE130 pKa = 4.22VRR132 pKa = 11.84NLQRR136 pKa = 11.84EE137 pKa = 4.34SNKK140 pKa = 10.13NVDD143 pKa = 3.46ILRR146 pKa = 11.84SMEE149 pKa = 4.01KK150 pKa = 9.8QGKK153 pKa = 5.94STEE156 pKa = 4.05MEE158 pKa = 3.51IAAAVATIRR167 pKa = 11.84SDD169 pKa = 2.9MVTAIKK175 pKa = 10.4SALKK179 pKa = 9.87EE180 pKa = 3.95QVTATSSSSGKK191 pKa = 9.92VEE193 pKa = 3.87LKK195 pKa = 10.81VRR197 pKa = 11.84TEE199 pKa = 3.86KK200 pKa = 10.71QAGLDD205 pKa = 3.69VKK207 pKa = 10.17QVPLLINPTTSKK219 pKa = 10.86LPRR222 pKa = 11.84KK223 pKa = 9.09PDD225 pKa = 3.45LEE227 pKa = 4.07AVRR230 pKa = 11.84PSSSKK235 pKa = 10.12SSKK238 pKa = 10.24EE239 pKa = 3.68VTLNEE244 pKa = 3.97LKK246 pKa = 10.54RR247 pKa = 11.84IFMCAVGVEE256 pKa = 3.86ADD258 pKa = 3.44IVSVLTDD265 pKa = 3.57EE266 pKa = 5.35EE267 pKa = 4.36VDD269 pKa = 4.66AMLTANDD276 pKa = 4.01YY277 pKa = 11.76YY278 pKa = 10.67EE279 pKa = 4.3FCNDD283 pKa = 3.76LDD285 pKa = 4.53DD286 pKa = 5.81DD287 pKa = 4.49GVRR290 pKa = 11.84KK291 pKa = 9.64EE292 pKa = 4.04FSEE295 pKa = 4.16IFNKK299 pKa = 10.91AIDD302 pKa = 3.93QLNKK306 pKa = 9.94EE307 pKa = 4.51AEE309 pKa = 4.26EE310 pKa = 4.1EE311 pKa = 4.22DD312 pKa = 3.65
Molecular weight: 34.49 kDa
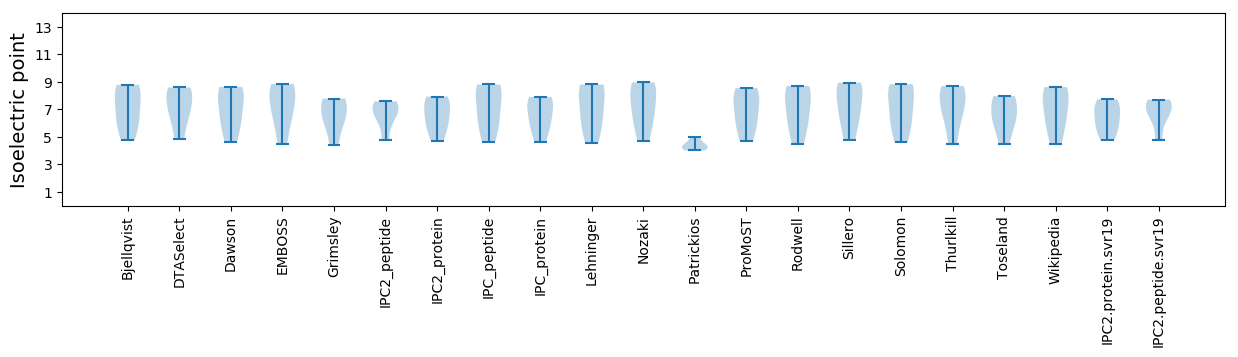
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R4WAR4|R4WAR4_9RHAB Matrix protein OS=Persimmon virus A OX=1211480 PE=4 SV=1
MM1 pKa = 7.34TSFVSTFKK9 pKa = 11.49GEE11 pKa = 4.08LNADD15 pKa = 3.48YY16 pKa = 8.79TQQVVKK22 pKa = 10.61LSKK25 pKa = 10.4KK26 pKa = 10.63LPILQSLWISAGFDD40 pKa = 3.07NVSITKK46 pKa = 10.09LAFSYY51 pKa = 10.71KK52 pKa = 9.97SRR54 pKa = 11.84CPKK57 pKa = 10.17AATGVVEE64 pKa = 3.55ITIRR68 pKa = 11.84DD69 pKa = 3.85LRR71 pKa = 11.84LEE73 pKa = 4.48DD74 pKa = 4.0ADD76 pKa = 3.79KK77 pKa = 11.54QEE79 pKa = 4.18VAFVTFNVKK88 pKa = 10.19DD89 pKa = 3.52WVEE92 pKa = 4.24LSWSYY97 pKa = 10.06PVWFHH102 pKa = 6.39CTDD105 pKa = 3.68FNGKK109 pKa = 8.95HH110 pKa = 5.51EE111 pKa = 4.81SVLDD115 pKa = 3.53MCLDD119 pKa = 3.74VVDD122 pKa = 4.84TNMSEE127 pKa = 4.05QFSLGSYY134 pKa = 9.46KK135 pKa = 9.93MKK137 pKa = 10.03IYY139 pKa = 10.81YY140 pKa = 9.68RR141 pKa = 11.84VQNQITKK148 pKa = 9.58FRR150 pKa = 11.84PSINRR155 pKa = 11.84AYY157 pKa = 10.84LIDD160 pKa = 3.64QTASPGAVITQRR172 pKa = 11.84RR173 pKa = 11.84KK174 pKa = 8.88EE175 pKa = 3.89DD176 pKa = 3.28HH177 pKa = 6.75NKK179 pKa = 9.92RR180 pKa = 11.84KK181 pKa = 10.19SNRR184 pKa = 11.84TSLSEE189 pKa = 4.02EE190 pKa = 4.07EE191 pKa = 4.29ISIQEE196 pKa = 3.72EE197 pKa = 4.79VIVHH201 pKa = 6.28KK202 pKa = 10.81EE203 pKa = 3.88NILDD207 pKa = 3.62RR208 pKa = 11.84SRR210 pKa = 11.84SYY212 pKa = 11.18LSSQPRR218 pKa = 11.84QCAKK222 pKa = 10.73
MM1 pKa = 7.34TSFVSTFKK9 pKa = 11.49GEE11 pKa = 4.08LNADD15 pKa = 3.48YY16 pKa = 8.79TQQVVKK22 pKa = 10.61LSKK25 pKa = 10.4KK26 pKa = 10.63LPILQSLWISAGFDD40 pKa = 3.07NVSITKK46 pKa = 10.09LAFSYY51 pKa = 10.71KK52 pKa = 9.97SRR54 pKa = 11.84CPKK57 pKa = 10.17AATGVVEE64 pKa = 3.55ITIRR68 pKa = 11.84DD69 pKa = 3.85LRR71 pKa = 11.84LEE73 pKa = 4.48DD74 pKa = 4.0ADD76 pKa = 3.79KK77 pKa = 11.54QEE79 pKa = 4.18VAFVTFNVKK88 pKa = 10.19DD89 pKa = 3.52WVEE92 pKa = 4.24LSWSYY97 pKa = 10.06PVWFHH102 pKa = 6.39CTDD105 pKa = 3.68FNGKK109 pKa = 8.95HH110 pKa = 5.51EE111 pKa = 4.81SVLDD115 pKa = 3.53MCLDD119 pKa = 3.74VVDD122 pKa = 4.84TNMSEE127 pKa = 4.05QFSLGSYY134 pKa = 9.46KK135 pKa = 9.93MKK137 pKa = 10.03IYY139 pKa = 10.81YY140 pKa = 9.68RR141 pKa = 11.84VQNQITKK148 pKa = 9.58FRR150 pKa = 11.84PSINRR155 pKa = 11.84AYY157 pKa = 10.84LIDD160 pKa = 3.64QTASPGAVITQRR172 pKa = 11.84RR173 pKa = 11.84KK174 pKa = 8.88EE175 pKa = 3.89DD176 pKa = 3.28HH177 pKa = 6.75NKK179 pKa = 9.92RR180 pKa = 11.84KK181 pKa = 10.19SNRR184 pKa = 11.84TSLSEE189 pKa = 4.02EE190 pKa = 4.07EE191 pKa = 4.29ISIQEE196 pKa = 3.72EE197 pKa = 4.79VIVHH201 pKa = 6.28KK202 pKa = 10.81EE203 pKa = 3.88NILDD207 pKa = 3.62RR208 pKa = 11.84SRR210 pKa = 11.84SYY212 pKa = 11.18LSSQPRR218 pKa = 11.84QCAKK222 pKa = 10.73
Molecular weight: 25.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3821 |
188 |
2089 |
636.8 |
72.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.711 ± 0.934 | 1.989 ± 0.401 |
6.281 ± 0.307 | 6.229 ± 0.596 |
3.298 ± 0.362 | 5.208 ± 0.416 |
1.937 ± 0.371 | 6.569 ± 0.531 |
7.485 ± 0.332 | 9.16 ± 0.515 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.083 ± 0.59 | 4.632 ± 0.759 |
4.057 ± 0.237 | 2.643 ± 0.377 |
5.025 ± 0.451 | 8.898 ± 0.632 |
6.386 ± 0.835 | 6.046 ± 0.974 |
1.439 ± 0.267 | 3.926 ± 0.544 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
