
Coemansia reversa (strain ATCC 12441 / NRRL 1564)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Zoopagomycota; Kickxellomycotina; Kickxellomycetes; Kickxellales; Kickxellaceae; Coemansia; Coemansia reversa
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
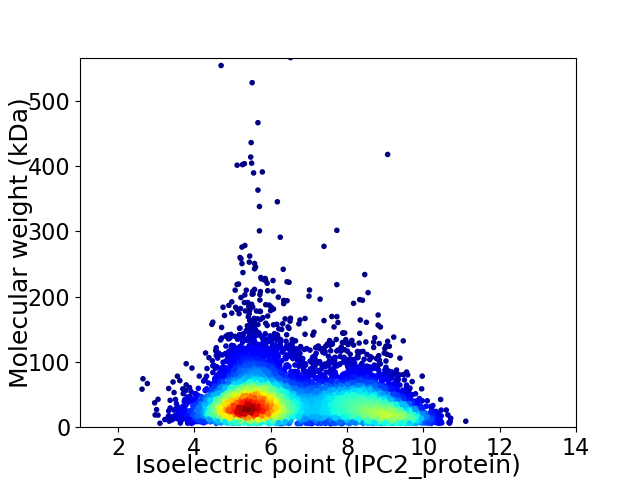
Virtual 2D-PAGE plot for 7317 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G5BIH1|A0A2G5BIH1_COERN Type I phosphodiesterase/nucleotide pyrophosphatase/phosphate transferase (Fragment) OS=Coemansia reversa (strain ATCC 12441 / NRRL 1564) OX=763665 GN=COEREDRAFT_22562 PE=4 SV=1
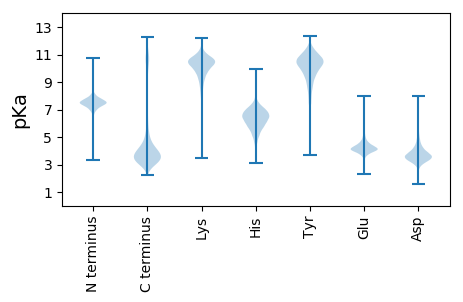
MM1 pKa = 7.71PRR3 pKa = 11.84PSDD6 pKa = 3.56FTDD9 pKa = 3.47CQARR13 pKa = 11.84DD14 pKa = 4.14DD15 pKa = 5.77EE16 pKa = 4.52IQCWIHH22 pKa = 6.23KK23 pKa = 9.14CHH25 pKa = 5.49EE26 pKa = 4.37HH27 pKa = 6.26EE28 pKa = 6.05DD29 pKa = 3.73IVGEE33 pKa = 4.07LTLDD37 pKa = 3.51FNKK40 pKa = 9.84VVTKK44 pKa = 10.51RR45 pKa = 11.84LHH47 pKa = 6.71RR48 pKa = 11.84DD49 pKa = 3.11VLYY52 pKa = 10.84HH53 pKa = 5.83LTNGHH58 pKa = 6.17GLGEE62 pKa = 4.19AAALDD67 pKa = 3.49AMVIVDD73 pKa = 3.63SASRR77 pKa = 11.84FLQVYY82 pKa = 8.38PIVNQTTATLIEE94 pKa = 4.34TLNSGWFLKK103 pKa = 11.27YY104 pKa = 10.54EE105 pKa = 4.68DD106 pKa = 4.87DD107 pKa = 4.05TNGMGWTKK115 pKa = 9.57FVPYY119 pKa = 9.91IVTSYY124 pKa = 11.16AVTVSEE130 pKa = 4.45SSSNIPATLFFGPSAIQSDD149 pKa = 4.32EE150 pKa = 4.17AFALQPTGGEE160 pKa = 4.27MPDD163 pKa = 3.32HH164 pKa = 5.8QQLMDD169 pKa = 4.02HH170 pKa = 5.88TWEE173 pKa = 4.95PYY175 pKa = 10.12HH176 pKa = 7.27VVARR180 pKa = 11.84YY181 pKa = 9.0GPSYY185 pKa = 10.42QLEE188 pKa = 4.34FADD191 pKa = 4.16GVPFVSTVAGDD202 pKa = 4.07LLMPYY207 pKa = 9.68HH208 pKa = 7.0WGDD211 pKa = 3.64DD212 pKa = 3.47VDD214 pKa = 4.13PAEE217 pKa = 4.45VAPSSTDD224 pKa = 3.06EE225 pKa = 4.6SDD227 pKa = 3.94PSDD230 pKa = 3.18HH231 pKa = 7.13SYY233 pKa = 11.51VDD235 pKa = 3.47EE236 pKa = 4.75SMNDD240 pKa = 3.61DD241 pKa = 3.66GSSNSSNAGPSTPPPEE257 pKa = 4.25EE258 pKa = 4.66SNGSLYY264 pKa = 9.76FTGSGGLGSSAALEE278 pKa = 4.33DD279 pKa = 3.44EE280 pKa = 4.69CRR282 pKa = 11.84RR283 pKa = 11.84SPSRR287 pKa = 11.84DD288 pKa = 2.98TSLQLPPSFLSFVAGRR304 pKa = 11.84DD305 pKa = 3.48TASQSPAHH313 pKa = 6.5ISVSSTNRR321 pKa = 11.84SLIRR325 pKa = 11.84GEE327 pKa = 4.03VPDD330 pKa = 3.72IVEE333 pKa = 4.26VLSDD337 pKa = 3.59SDD339 pKa = 4.03EE340 pKa = 4.4SDD342 pKa = 4.24NYY344 pKa = 11.45DD345 pKa = 3.39SDD347 pKa = 4.04DD348 pKa = 3.91SEE350 pKa = 4.64YY351 pKa = 11.25EE352 pKa = 4.02PLSGDD357 pKa = 3.35NTSDD361 pKa = 3.37KK362 pKa = 11.38SSDD365 pKa = 3.41EE366 pKa = 4.18SDD368 pKa = 4.09GKK370 pKa = 10.71DD371 pKa = 3.08EE372 pKa = 5.8DD373 pKa = 4.51EE374 pKa = 4.49GTEE377 pKa = 4.1PPLGPLAVSSIGFSGPSSIRR397 pKa = 11.84NATTMSEE404 pKa = 4.11MEE406 pKa = 4.61TIQSTMVCDD415 pKa = 4.21ISTAPNTCDD424 pKa = 3.05VGVSTEE430 pKa = 3.8NMYY433 pKa = 8.12TTASAQTNTASDD445 pKa = 3.82MLEE448 pKa = 3.58QAQYY452 pKa = 11.76GEE454 pKa = 4.54TPTDD458 pKa = 3.22NATMVGIEE466 pKa = 4.58TIMVDD471 pKa = 4.52DD472 pKa = 5.49GDD474 pKa = 4.89SISNDD479 pKa = 3.57EE480 pKa = 3.77IRR482 pKa = 11.84IDD484 pKa = 3.37IVEE487 pKa = 4.58DD488 pKa = 3.8EE489 pKa = 4.29EE490 pKa = 5.55DD491 pKa = 3.54SGRR494 pKa = 11.84SAIVPLPQVPSPQTVTTKK512 pKa = 10.97APDD515 pKa = 3.37NHH517 pKa = 7.09IDD519 pKa = 3.59AEE521 pKa = 4.55TIPTSDD527 pKa = 3.78NSPSEE532 pKa = 3.67IWAMSTAIVPYY543 pKa = 8.33SHH545 pKa = 7.01GYY547 pKa = 8.71YY548 pKa = 10.37GYY550 pKa = 10.5SPYY553 pKa = 10.93EE554 pKa = 3.88LAVSRR559 pKa = 11.84ALINN563 pKa = 3.45
MM1 pKa = 7.71PRR3 pKa = 11.84PSDD6 pKa = 3.56FTDD9 pKa = 3.47CQARR13 pKa = 11.84DD14 pKa = 4.14DD15 pKa = 5.77EE16 pKa = 4.52IQCWIHH22 pKa = 6.23KK23 pKa = 9.14CHH25 pKa = 5.49EE26 pKa = 4.37HH27 pKa = 6.26EE28 pKa = 6.05DD29 pKa = 3.73IVGEE33 pKa = 4.07LTLDD37 pKa = 3.51FNKK40 pKa = 9.84VVTKK44 pKa = 10.51RR45 pKa = 11.84LHH47 pKa = 6.71RR48 pKa = 11.84DD49 pKa = 3.11VLYY52 pKa = 10.84HH53 pKa = 5.83LTNGHH58 pKa = 6.17GLGEE62 pKa = 4.19AAALDD67 pKa = 3.49AMVIVDD73 pKa = 3.63SASRR77 pKa = 11.84FLQVYY82 pKa = 8.38PIVNQTTATLIEE94 pKa = 4.34TLNSGWFLKK103 pKa = 11.27YY104 pKa = 10.54EE105 pKa = 4.68DD106 pKa = 4.87DD107 pKa = 4.05TNGMGWTKK115 pKa = 9.57FVPYY119 pKa = 9.91IVTSYY124 pKa = 11.16AVTVSEE130 pKa = 4.45SSSNIPATLFFGPSAIQSDD149 pKa = 4.32EE150 pKa = 4.17AFALQPTGGEE160 pKa = 4.27MPDD163 pKa = 3.32HH164 pKa = 5.8QQLMDD169 pKa = 4.02HH170 pKa = 5.88TWEE173 pKa = 4.95PYY175 pKa = 10.12HH176 pKa = 7.27VVARR180 pKa = 11.84YY181 pKa = 9.0GPSYY185 pKa = 10.42QLEE188 pKa = 4.34FADD191 pKa = 4.16GVPFVSTVAGDD202 pKa = 4.07LLMPYY207 pKa = 9.68HH208 pKa = 7.0WGDD211 pKa = 3.64DD212 pKa = 3.47VDD214 pKa = 4.13PAEE217 pKa = 4.45VAPSSTDD224 pKa = 3.06EE225 pKa = 4.6SDD227 pKa = 3.94PSDD230 pKa = 3.18HH231 pKa = 7.13SYY233 pKa = 11.51VDD235 pKa = 3.47EE236 pKa = 4.75SMNDD240 pKa = 3.61DD241 pKa = 3.66GSSNSSNAGPSTPPPEE257 pKa = 4.25EE258 pKa = 4.66SNGSLYY264 pKa = 9.76FTGSGGLGSSAALEE278 pKa = 4.33DD279 pKa = 3.44EE280 pKa = 4.69CRR282 pKa = 11.84RR283 pKa = 11.84SPSRR287 pKa = 11.84DD288 pKa = 2.98TSLQLPPSFLSFVAGRR304 pKa = 11.84DD305 pKa = 3.48TASQSPAHH313 pKa = 6.5ISVSSTNRR321 pKa = 11.84SLIRR325 pKa = 11.84GEE327 pKa = 4.03VPDD330 pKa = 3.72IVEE333 pKa = 4.26VLSDD337 pKa = 3.59SDD339 pKa = 4.03EE340 pKa = 4.4SDD342 pKa = 4.24NYY344 pKa = 11.45DD345 pKa = 3.39SDD347 pKa = 4.04DD348 pKa = 3.91SEE350 pKa = 4.64YY351 pKa = 11.25EE352 pKa = 4.02PLSGDD357 pKa = 3.35NTSDD361 pKa = 3.37KK362 pKa = 11.38SSDD365 pKa = 3.41EE366 pKa = 4.18SDD368 pKa = 4.09GKK370 pKa = 10.71DD371 pKa = 3.08EE372 pKa = 5.8DD373 pKa = 4.51EE374 pKa = 4.49GTEE377 pKa = 4.1PPLGPLAVSSIGFSGPSSIRR397 pKa = 11.84NATTMSEE404 pKa = 4.11MEE406 pKa = 4.61TIQSTMVCDD415 pKa = 4.21ISTAPNTCDD424 pKa = 3.05VGVSTEE430 pKa = 3.8NMYY433 pKa = 8.12TTASAQTNTASDD445 pKa = 3.82MLEE448 pKa = 3.58QAQYY452 pKa = 11.76GEE454 pKa = 4.54TPTDD458 pKa = 3.22NATMVGIEE466 pKa = 4.58TIMVDD471 pKa = 4.52DD472 pKa = 5.49GDD474 pKa = 4.89SISNDD479 pKa = 3.57EE480 pKa = 3.77IRR482 pKa = 11.84IDD484 pKa = 3.37IVEE487 pKa = 4.58DD488 pKa = 3.8EE489 pKa = 4.29EE490 pKa = 5.55DD491 pKa = 3.54SGRR494 pKa = 11.84SAIVPLPQVPSPQTVTTKK512 pKa = 10.97APDD515 pKa = 3.37NHH517 pKa = 7.09IDD519 pKa = 3.59AEE521 pKa = 4.55TIPTSDD527 pKa = 3.78NSPSEE532 pKa = 3.67IWAMSTAIVPYY543 pKa = 8.33SHH545 pKa = 7.01GYY547 pKa = 8.71YY548 pKa = 10.37GYY550 pKa = 10.5SPYY553 pKa = 10.93EE554 pKa = 3.88LAVSRR559 pKa = 11.84ALINN563 pKa = 3.45
Molecular weight: 60.81 kDa
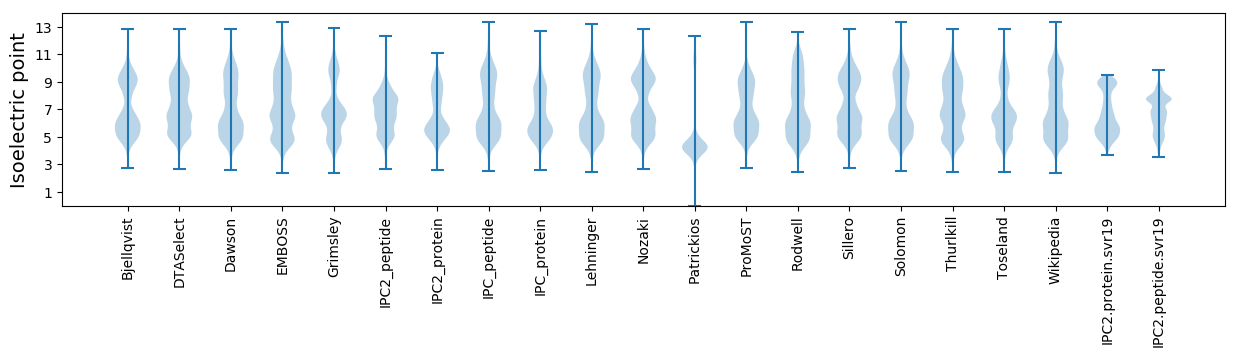
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G5B6P2|A0A2G5B6P2_COERN Uncharacterized protein OS=Coemansia reversa (strain ATCC 12441 / NRRL 1564) OX=763665 GN=COEREDRAFT_82558 PE=4 SV=1
MM1 pKa = 6.53TTKK4 pKa = 10.69LPPGPRR10 pKa = 11.84MMTTKK15 pKa = 10.59LPPGPRR21 pKa = 11.84MMTTKK26 pKa = 10.59LPPGPRR32 pKa = 11.84MMTTKK37 pKa = 10.59LPPGPRR43 pKa = 11.84MATTRR48 pKa = 11.84LPPGLRR54 pKa = 11.84MVTTKK59 pKa = 10.76LPPGRR64 pKa = 11.84MTVAMPVPPGPRR76 pKa = 11.84MMTTKK81 pKa = 10.76LPPP84 pKa = 4.06
MM1 pKa = 6.53TTKK4 pKa = 10.69LPPGPRR10 pKa = 11.84MMTTKK15 pKa = 10.59LPPGPRR21 pKa = 11.84MMTTKK26 pKa = 10.59LPPGPRR32 pKa = 11.84MMTTKK37 pKa = 10.59LPPGPRR43 pKa = 11.84MATTRR48 pKa = 11.84LPPGLRR54 pKa = 11.84MVTTKK59 pKa = 10.76LPPGRR64 pKa = 11.84MTVAMPVPPGPRR76 pKa = 11.84MMTTKK81 pKa = 10.76LPPP84 pKa = 4.06
Molecular weight: 9.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2997087 |
49 |
5170 |
409.6 |
45.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.114 ± 0.043 | 1.461 ± 0.013 |
5.695 ± 0.022 | 5.879 ± 0.026 |
3.525 ± 0.018 | 6.286 ± 0.023 |
2.524 ± 0.014 | 5.13 ± 0.025 |
4.502 ± 0.026 | 9.115 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.492 ± 0.012 | 3.988 ± 0.022 |
5.001 ± 0.028 | 4.279 ± 0.023 |
6.272 ± 0.03 | 8.233 ± 0.032 |
5.816 ± 0.019 | 6.62 ± 0.023 |
1.17 ± 0.01 | 2.899 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
