
Pseudomonas sp. BAY1663
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; unclassified Pseudomonas
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
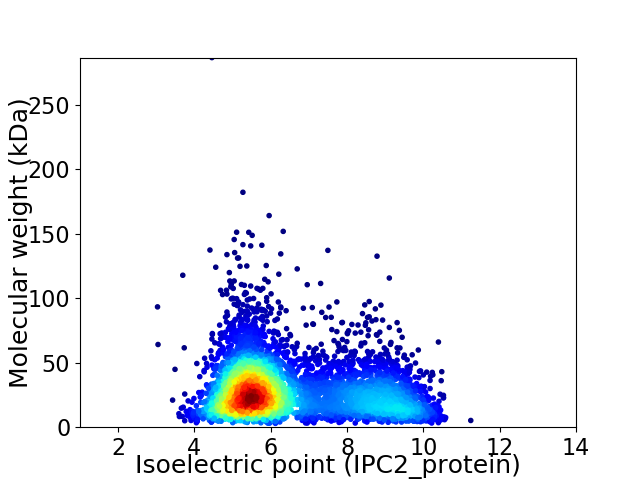
Virtual 2D-PAGE plot for 5053 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W9SY05|W9SY05_9PSED Uncharacterized protein OS=Pseudomonas sp. BAY1663 OX=1439940 GN=BAY1663_04462 PE=4 SV=1
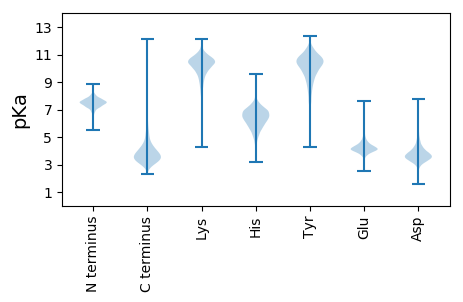
MM1 pKa = 7.87ASTLDD6 pKa = 3.39ISNEE10 pKa = 3.49DD11 pKa = 3.51FAAGNYY17 pKa = 8.82RR18 pKa = 11.84FKK20 pKa = 11.35GDD22 pKa = 3.37GSNASVINQGTLMASDD38 pKa = 4.31GGAVALLGGTVSNQGVIVANSGTVALAAGNAVTLDD73 pKa = 3.62FAGDD77 pKa = 3.48GLLNVQVDD85 pKa = 4.3EE86 pKa = 4.31ATKK89 pKa = 10.72DD90 pKa = 3.37ALVEE94 pKa = 3.85NRR96 pKa = 11.84QLIQADD102 pKa = 4.06GGQVLLTAQAADD114 pKa = 3.25ALLRR118 pKa = 11.84TVVNNDD124 pKa = 3.7GIIEE128 pKa = 4.11ARR130 pKa = 11.84TLGEE134 pKa = 4.06KK135 pKa = 9.92NGKK138 pKa = 9.11VVLNGGDD145 pKa = 3.37EE146 pKa = 4.46GSVQVAGTLDD156 pKa = 3.47TSATTGQGGAIEE168 pKa = 4.16VTGANVALADD178 pKa = 3.78ATLDD182 pKa = 3.45ASGATGGGSVKK193 pKa = 10.73VGGDD197 pKa = 3.22WQGTGTLTRR206 pKa = 11.84AQATTVDD213 pKa = 3.41AASTLKK219 pKa = 10.73ADD221 pKa = 3.72ATHH224 pKa = 6.61NGDD227 pKa = 3.14GGTVVVWSDD236 pKa = 2.94QHH238 pKa = 5.8TSYY241 pKa = 10.49QGHH244 pKa = 6.82ISAKK248 pKa = 10.26GGAQGGDD255 pKa = 3.0GGQAEE260 pKa = 4.57VSGKK264 pKa = 10.45AVLGFTGTVDD274 pKa = 3.78LSAAHH279 pKa = 6.53GAFGDD284 pKa = 4.27LLLDD288 pKa = 4.72PYY290 pKa = 11.32DD291 pKa = 3.79LTISSGTGGSATATADD307 pKa = 4.09DD308 pKa = 4.21STLGADD314 pKa = 3.59TLEE317 pKa = 4.5GLLADD322 pKa = 4.29ANVTVSTGTAGSQNGDD338 pKa = 2.95ITVASAVSWDD348 pKa = 3.59ANTTLTLNAAGNIDD362 pKa = 3.51IDD364 pKa = 3.98AAITASGEE372 pKa = 4.41SAGLALNYY380 pKa = 10.59GGTEE384 pKa = 4.2GVAGATAASGTDD396 pKa = 3.55YY397 pKa = 11.25NVNAPVTLSGADD409 pKa = 3.1ATLSINGSDD418 pKa = 3.51YY419 pKa = 11.3TLIHH423 pKa = 7.68SMEE426 pKa = 4.1QLDD429 pKa = 5.13AIDD432 pKa = 3.67STGLSGHH439 pKa = 6.0YY440 pKa = 10.54ALAQDD445 pKa = 5.18LDD447 pKa = 4.45ASDD450 pKa = 4.28SPYY453 pKa = 10.72TSALVGIDD461 pKa = 3.25AANAFTGTFAGLGHH475 pKa = 7.71AIGNLTISASGSDD488 pKa = 3.82YY489 pKa = 11.49VGLFGYY495 pKa = 8.39TGSGSTLRR503 pKa = 11.84DD504 pKa = 3.03VGLQGGSVTGNSYY517 pKa = 10.94VGGLVGYY524 pKa = 9.93NDD526 pKa = 4.03GGSIVDD532 pKa = 4.37AYY534 pKa = 10.52ATVDD538 pKa = 3.18VTGAHH543 pKa = 4.55VTGGLVGANVGGGSILRR560 pKa = 11.84AYY562 pKa = 7.99ATGWVNATASEE573 pKa = 4.19GLGWAGGLIGQNAGSISQSYY593 pKa = 10.58ASGGVTGDD601 pKa = 3.71YY602 pKa = 10.88YY603 pKa = 11.47VGGLVGHH610 pKa = 6.77NGNGTVTDD618 pKa = 4.47SYY620 pKa = 12.07ALGNVHH626 pKa = 6.12GQWDD630 pKa = 3.67VGGG633 pKa = 3.82
MM1 pKa = 7.87ASTLDD6 pKa = 3.39ISNEE10 pKa = 3.49DD11 pKa = 3.51FAAGNYY17 pKa = 8.82RR18 pKa = 11.84FKK20 pKa = 11.35GDD22 pKa = 3.37GSNASVINQGTLMASDD38 pKa = 4.31GGAVALLGGTVSNQGVIVANSGTVALAAGNAVTLDD73 pKa = 3.62FAGDD77 pKa = 3.48GLLNVQVDD85 pKa = 4.3EE86 pKa = 4.31ATKK89 pKa = 10.72DD90 pKa = 3.37ALVEE94 pKa = 3.85NRR96 pKa = 11.84QLIQADD102 pKa = 4.06GGQVLLTAQAADD114 pKa = 3.25ALLRR118 pKa = 11.84TVVNNDD124 pKa = 3.7GIIEE128 pKa = 4.11ARR130 pKa = 11.84TLGEE134 pKa = 4.06KK135 pKa = 9.92NGKK138 pKa = 9.11VVLNGGDD145 pKa = 3.37EE146 pKa = 4.46GSVQVAGTLDD156 pKa = 3.47TSATTGQGGAIEE168 pKa = 4.16VTGANVALADD178 pKa = 3.78ATLDD182 pKa = 3.45ASGATGGGSVKK193 pKa = 10.73VGGDD197 pKa = 3.22WQGTGTLTRR206 pKa = 11.84AQATTVDD213 pKa = 3.41AASTLKK219 pKa = 10.73ADD221 pKa = 3.72ATHH224 pKa = 6.61NGDD227 pKa = 3.14GGTVVVWSDD236 pKa = 2.94QHH238 pKa = 5.8TSYY241 pKa = 10.49QGHH244 pKa = 6.82ISAKK248 pKa = 10.26GGAQGGDD255 pKa = 3.0GGQAEE260 pKa = 4.57VSGKK264 pKa = 10.45AVLGFTGTVDD274 pKa = 3.78LSAAHH279 pKa = 6.53GAFGDD284 pKa = 4.27LLLDD288 pKa = 4.72PYY290 pKa = 11.32DD291 pKa = 3.79LTISSGTGGSATATADD307 pKa = 4.09DD308 pKa = 4.21STLGADD314 pKa = 3.59TLEE317 pKa = 4.5GLLADD322 pKa = 4.29ANVTVSTGTAGSQNGDD338 pKa = 2.95ITVASAVSWDD348 pKa = 3.59ANTTLTLNAAGNIDD362 pKa = 3.51IDD364 pKa = 3.98AAITASGEE372 pKa = 4.41SAGLALNYY380 pKa = 10.59GGTEE384 pKa = 4.2GVAGATAASGTDD396 pKa = 3.55YY397 pKa = 11.25NVNAPVTLSGADD409 pKa = 3.1ATLSINGSDD418 pKa = 3.51YY419 pKa = 11.3TLIHH423 pKa = 7.68SMEE426 pKa = 4.1QLDD429 pKa = 5.13AIDD432 pKa = 3.67STGLSGHH439 pKa = 6.0YY440 pKa = 10.54ALAQDD445 pKa = 5.18LDD447 pKa = 4.45ASDD450 pKa = 4.28SPYY453 pKa = 10.72TSALVGIDD461 pKa = 3.25AANAFTGTFAGLGHH475 pKa = 7.71AIGNLTISASGSDD488 pKa = 3.82YY489 pKa = 11.49VGLFGYY495 pKa = 8.39TGSGSTLRR503 pKa = 11.84DD504 pKa = 3.03VGLQGGSVTGNSYY517 pKa = 10.94VGGLVGYY524 pKa = 9.93NDD526 pKa = 4.03GGSIVDD532 pKa = 4.37AYY534 pKa = 10.52ATVDD538 pKa = 3.18VTGAHH543 pKa = 4.55VTGGLVGANVGGGSILRR560 pKa = 11.84AYY562 pKa = 7.99ATGWVNATASEE573 pKa = 4.19GLGWAGGLIGQNAGSISQSYY593 pKa = 10.58ASGGVTGDD601 pKa = 3.71YY602 pKa = 10.88YY603 pKa = 11.47VGGLVGHH610 pKa = 6.77NGNGTVTDD618 pKa = 4.47SYY620 pKa = 12.07ALGNVHH626 pKa = 6.12GQWDD630 pKa = 3.67VGGG633 pKa = 3.82
Molecular weight: 61.53 kDa
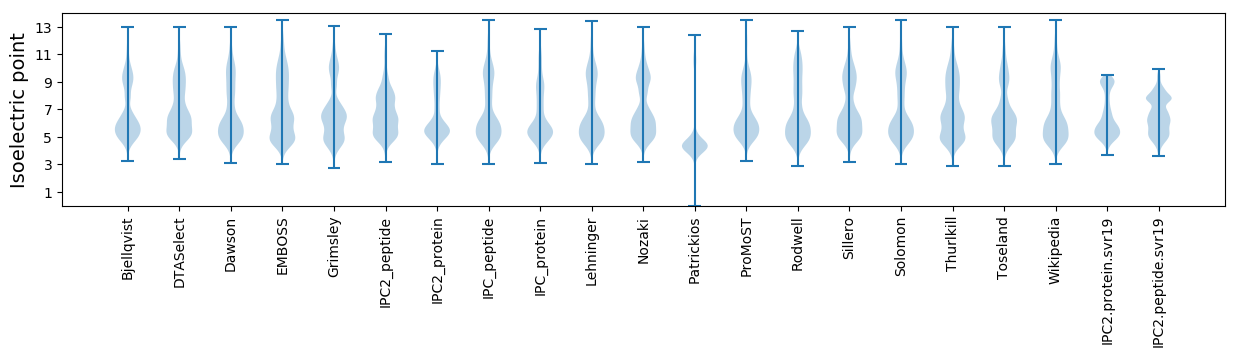
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W9T8H7|W9T8H7_9PSED Two-component response regulator OS=Pseudomonas sp. BAY1663 OX=1439940 GN=BAY1663_03887 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1391436 |
29 |
2815 |
275.4 |
30.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.504 ± 0.051 | 1.09 ± 0.013 |
5.295 ± 0.025 | 6.091 ± 0.036 |
3.486 ± 0.021 | 8.104 ± 0.034 |
2.302 ± 0.019 | 4.603 ± 0.025 |
3.046 ± 0.031 | 11.939 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.305 ± 0.018 | 2.69 ± 0.022 |
5.008 ± 0.027 | 4.475 ± 0.033 |
7.327 ± 0.036 | 5.522 ± 0.027 |
4.519 ± 0.026 | 6.813 ± 0.032 |
1.452 ± 0.016 | 2.428 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
