
Natronospirillum operosum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Natronospirillaceae; Natronospirillum
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
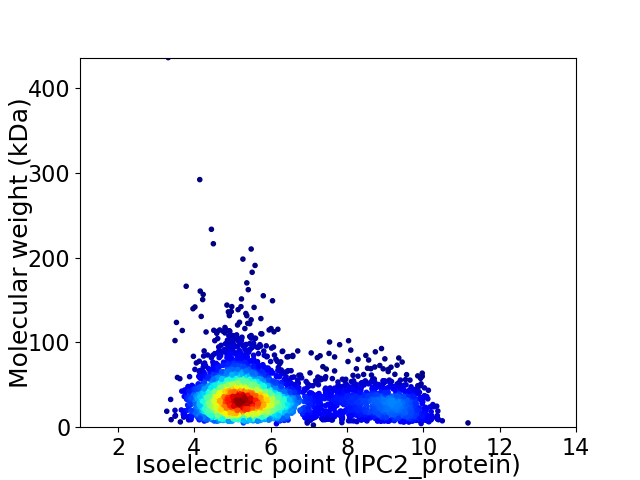
Virtual 2D-PAGE plot for 3948 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
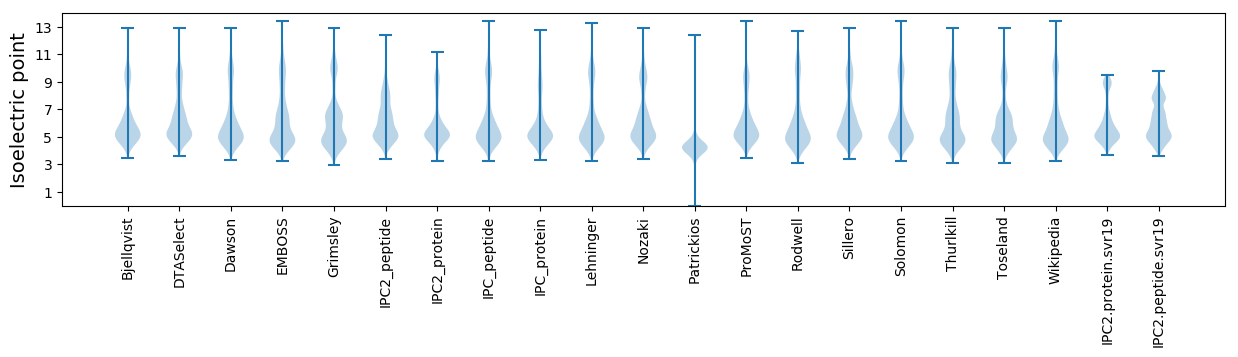
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Z0WC45|A0A4Z0WC45_9GAMM DMT family transporter OS=Natronospirillum operosum OX=2759953 GN=E4656_15490 PE=4 SV=1
MM1 pKa = 7.4SPCTHH6 pKa = 6.74KK7 pKa = 10.75PLLSHH12 pKa = 7.04LLVLLAVTALAACNPSTEE30 pKa = 4.48SGGNSDD36 pKa = 4.72NGNGDD41 pKa = 3.3GGGGNGGNDD50 pKa = 3.28NNGDD54 pKa = 4.11LVEE57 pKa = 4.8LDD59 pKa = 3.69YY60 pKa = 11.69GVGFVLVMDD69 pKa = 5.09PAPNQIALRR78 pKa = 11.84GYY80 pKa = 8.46GTGQIPEE87 pKa = 4.28TTRR90 pKa = 11.84LAVQVLDD97 pKa = 3.85NDD99 pKa = 4.24LNPVAGHH106 pKa = 6.65DD107 pKa = 3.54VSFAITDD114 pKa = 3.64STGGPEE120 pKa = 4.12LTRR123 pKa = 11.84DD124 pKa = 3.49SATTGSDD131 pKa = 3.11GSASTILRR139 pKa = 11.84SGTVQGTTQVEE150 pKa = 4.34VSMDD154 pKa = 3.7LLDD157 pKa = 4.15SNGNVIEE164 pKa = 4.04QDD166 pKa = 3.34VHH168 pKa = 6.47VATTQPVSMHH178 pKa = 5.78TAAPVTDD185 pKa = 3.88GMSLSADD192 pKa = 3.44VFNPEE197 pKa = 3.96GYY199 pKa = 9.72DD200 pKa = 3.47YY201 pKa = 11.61NGIPVQVTVHH211 pKa = 6.48LGDD214 pKa = 3.21IHH216 pKa = 7.18RR217 pKa = 11.84NPVADD222 pKa = 3.71GTQVNFLASTGLVDD236 pKa = 5.37PVCTTDD242 pKa = 5.03NGACTIEE249 pKa = 4.2WRR251 pKa = 11.84SAHH254 pKa = 6.05PRR256 pKa = 11.84GDD258 pKa = 3.14GRR260 pKa = 11.84VDD262 pKa = 2.9ILARR266 pKa = 11.84TNGNDD271 pKa = 3.72AFVDD275 pKa = 3.7TNGNSRR281 pKa = 11.84FDD283 pKa = 3.54LGEE286 pKa = 3.97SYY288 pKa = 10.56EE289 pKa = 4.21PQGEE293 pKa = 4.16PFVDD297 pKa = 3.87TEE299 pKa = 4.43GTGQFDD305 pKa = 3.64PGDD308 pKa = 4.08FFWDD312 pKa = 3.99LSGTGDD318 pKa = 3.97YY319 pKa = 10.66TPADD323 pKa = 3.89PGAHH327 pKa = 5.68YY328 pKa = 10.36KK329 pKa = 10.99GIGCTEE335 pKa = 4.19DD336 pKa = 3.69ALEE339 pKa = 5.16HH340 pKa = 5.23GHH342 pKa = 6.3CDD344 pKa = 2.97EE345 pKa = 4.98RR346 pKa = 11.84AVVFNNIVLVASGSDD361 pKa = 3.29LRR363 pKa = 11.84LDD365 pKa = 3.99PEE367 pKa = 4.32PTDD370 pKa = 3.45VSVSDD375 pKa = 3.35TVIYY379 pKa = 10.74QMVDD383 pKa = 2.85QNGNVPPAGSDD394 pKa = 3.42LSINCTDD401 pKa = 3.88ASATALDD408 pKa = 3.94SEE410 pKa = 4.88VPEE413 pKa = 4.27RR414 pKa = 11.84PGALFGPWPVVINYY428 pKa = 9.96GDD430 pKa = 4.21DD431 pKa = 3.8PGSCTLEE438 pKa = 3.95VQSPNGLPQRR448 pKa = 11.84YY449 pKa = 6.73TISVNN454 pKa = 3.15
MM1 pKa = 7.4SPCTHH6 pKa = 6.74KK7 pKa = 10.75PLLSHH12 pKa = 7.04LLVLLAVTALAACNPSTEE30 pKa = 4.48SGGNSDD36 pKa = 4.72NGNGDD41 pKa = 3.3GGGGNGGNDD50 pKa = 3.28NNGDD54 pKa = 4.11LVEE57 pKa = 4.8LDD59 pKa = 3.69YY60 pKa = 11.69GVGFVLVMDD69 pKa = 5.09PAPNQIALRR78 pKa = 11.84GYY80 pKa = 8.46GTGQIPEE87 pKa = 4.28TTRR90 pKa = 11.84LAVQVLDD97 pKa = 3.85NDD99 pKa = 4.24LNPVAGHH106 pKa = 6.65DD107 pKa = 3.54VSFAITDD114 pKa = 3.64STGGPEE120 pKa = 4.12LTRR123 pKa = 11.84DD124 pKa = 3.49SATTGSDD131 pKa = 3.11GSASTILRR139 pKa = 11.84SGTVQGTTQVEE150 pKa = 4.34VSMDD154 pKa = 3.7LLDD157 pKa = 4.15SNGNVIEE164 pKa = 4.04QDD166 pKa = 3.34VHH168 pKa = 6.47VATTQPVSMHH178 pKa = 5.78TAAPVTDD185 pKa = 3.88GMSLSADD192 pKa = 3.44VFNPEE197 pKa = 3.96GYY199 pKa = 9.72DD200 pKa = 3.47YY201 pKa = 11.61NGIPVQVTVHH211 pKa = 6.48LGDD214 pKa = 3.21IHH216 pKa = 7.18RR217 pKa = 11.84NPVADD222 pKa = 3.71GTQVNFLASTGLVDD236 pKa = 5.37PVCTTDD242 pKa = 5.03NGACTIEE249 pKa = 4.2WRR251 pKa = 11.84SAHH254 pKa = 6.05PRR256 pKa = 11.84GDD258 pKa = 3.14GRR260 pKa = 11.84VDD262 pKa = 2.9ILARR266 pKa = 11.84TNGNDD271 pKa = 3.72AFVDD275 pKa = 3.7TNGNSRR281 pKa = 11.84FDD283 pKa = 3.54LGEE286 pKa = 3.97SYY288 pKa = 10.56EE289 pKa = 4.21PQGEE293 pKa = 4.16PFVDD297 pKa = 3.87TEE299 pKa = 4.43GTGQFDD305 pKa = 3.64PGDD308 pKa = 4.08FFWDD312 pKa = 3.99LSGTGDD318 pKa = 3.97YY319 pKa = 10.66TPADD323 pKa = 3.89PGAHH327 pKa = 5.68YY328 pKa = 10.36KK329 pKa = 10.99GIGCTEE335 pKa = 4.19DD336 pKa = 3.69ALEE339 pKa = 5.16HH340 pKa = 5.23GHH342 pKa = 6.3CDD344 pKa = 2.97EE345 pKa = 4.98RR346 pKa = 11.84AVVFNNIVLVASGSDD361 pKa = 3.29LRR363 pKa = 11.84LDD365 pKa = 3.99PEE367 pKa = 4.32PTDD370 pKa = 3.45VSVSDD375 pKa = 3.35TVIYY379 pKa = 10.74QMVDD383 pKa = 2.85QNGNVPPAGSDD394 pKa = 3.42LSINCTDD401 pKa = 3.88ASATALDD408 pKa = 3.94SEE410 pKa = 4.88VPEE413 pKa = 4.27RR414 pKa = 11.84PGALFGPWPVVINYY428 pKa = 9.96GDD430 pKa = 4.21DD431 pKa = 3.8PGSCTLEE438 pKa = 3.95VQSPNGLPQRR448 pKa = 11.84YY449 pKa = 6.73TISVNN454 pKa = 3.15
Molecular weight: 47.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Z0WBI7|A0A4Z0WBI7_9GAMM Beta-N-acetylhexosaminidase OS=Natronospirillum operosum OX=2759953 GN=E4656_06875 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 8.98ILAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.72GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 8.98ILAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.72GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1311596 |
22 |
4272 |
332.2 |
36.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.46 ± 0.042 | 0.879 ± 0.013 |
5.926 ± 0.039 | 6.186 ± 0.042 |
3.553 ± 0.027 | 7.659 ± 0.041 |
2.423 ± 0.021 | 4.934 ± 0.033 |
2.456 ± 0.035 | 11.364 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.558 ± 0.022 | 2.893 ± 0.028 |
4.852 ± 0.032 | 4.688 ± 0.028 |
6.985 ± 0.039 | 5.617 ± 0.026 |
5.34 ± 0.04 | 7.127 ± 0.03 |
1.584 ± 0.019 | 2.515 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
