
Circoviridae 6 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
Average proteome isoelectric point is 8.1
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
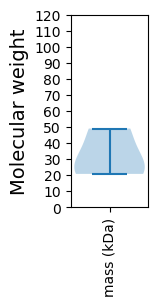
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|S5TNC2|S5TNC2_9CIRC Replication-associated protein OS=Circoviridae 6 LDMD-2013 OX=1379710 PE=4 SV=1
MM1 pKa = 7.68SDD3 pKa = 3.25VPAAPPSTVAAGRR16 pKa = 11.84SRR18 pKa = 11.84GWTGSMFDD26 pKa = 3.62YY27 pKa = 11.13DD28 pKa = 4.92DD29 pKa = 4.35VYY31 pKa = 11.59EE32 pKa = 4.43SLCEE36 pKa = 3.79ALAAAVSEE44 pKa = 4.11QRR46 pKa = 11.84VTRR49 pKa = 11.84AILGKK54 pKa = 8.23EE55 pKa = 4.06VCPTTQRR62 pKa = 11.84KK63 pKa = 7.77HH64 pKa = 5.05LQWYY68 pKa = 9.29VEE70 pKa = 4.37FGNAVQRR77 pKa = 11.84GSVTNLGITGHH88 pKa = 5.25TEE90 pKa = 3.42HH91 pKa = 7.51RR92 pKa = 11.84LGSPAQCWQYY102 pKa = 11.05CSKK105 pKa = 11.11DD106 pKa = 2.99GDD108 pKa = 3.85FATFGSPPKK117 pKa = 10.4GQGKK121 pKa = 10.15RR122 pKa = 11.84SDD124 pKa = 3.6LDD126 pKa = 3.41QAIEE130 pKa = 4.11TLTSEE135 pKa = 4.32GLEE138 pKa = 4.1ATAMEE143 pKa = 4.68HH144 pKa = 6.05PAAFVKK150 pKa = 10.62YY151 pKa = 10.04GRR153 pKa = 11.84GLAALEE159 pKa = 4.05ARR161 pKa = 11.84FTRR164 pKa = 11.84PFPLPQPTSPPRR176 pKa = 11.84YY177 pKa = 6.68VTPVRR182 pKa = 11.84VRR184 pKa = 11.84LAEE187 pKa = 3.95YY188 pKa = 8.92PQEE191 pKa = 5.51AEE193 pKa = 4.66APPPTPRR200 pKa = 11.84RR201 pKa = 11.84HH202 pKa = 5.68GGSGRR207 pKa = 11.84GCVVGPDD214 pKa = 4.15GNWEE218 pKa = 3.83IPYY221 pKa = 9.58GVPGGIGSVCMGTRR235 pKa = 11.84DD236 pKa = 3.28GSMVV240 pKa = 3.03
MM1 pKa = 7.68SDD3 pKa = 3.25VPAAPPSTVAAGRR16 pKa = 11.84SRR18 pKa = 11.84GWTGSMFDD26 pKa = 3.62YY27 pKa = 11.13DD28 pKa = 4.92DD29 pKa = 4.35VYY31 pKa = 11.59EE32 pKa = 4.43SLCEE36 pKa = 3.79ALAAAVSEE44 pKa = 4.11QRR46 pKa = 11.84VTRR49 pKa = 11.84AILGKK54 pKa = 8.23EE55 pKa = 4.06VCPTTQRR62 pKa = 11.84KK63 pKa = 7.77HH64 pKa = 5.05LQWYY68 pKa = 9.29VEE70 pKa = 4.37FGNAVQRR77 pKa = 11.84GSVTNLGITGHH88 pKa = 5.25TEE90 pKa = 3.42HH91 pKa = 7.51RR92 pKa = 11.84LGSPAQCWQYY102 pKa = 11.05CSKK105 pKa = 11.11DD106 pKa = 2.99GDD108 pKa = 3.85FATFGSPPKK117 pKa = 10.4GQGKK121 pKa = 10.15RR122 pKa = 11.84SDD124 pKa = 3.6LDD126 pKa = 3.41QAIEE130 pKa = 4.11TLTSEE135 pKa = 4.32GLEE138 pKa = 4.1ATAMEE143 pKa = 4.68HH144 pKa = 6.05PAAFVKK150 pKa = 10.62YY151 pKa = 10.04GRR153 pKa = 11.84GLAALEE159 pKa = 4.05ARR161 pKa = 11.84FTRR164 pKa = 11.84PFPLPQPTSPPRR176 pKa = 11.84YY177 pKa = 6.68VTPVRR182 pKa = 11.84VRR184 pKa = 11.84LAEE187 pKa = 3.95YY188 pKa = 8.92PQEE191 pKa = 5.51AEE193 pKa = 4.66APPPTPRR200 pKa = 11.84RR201 pKa = 11.84HH202 pKa = 5.68GGSGRR207 pKa = 11.84GCVVGPDD214 pKa = 4.15GNWEE218 pKa = 3.83IPYY221 pKa = 9.58GVPGGIGSVCMGTRR235 pKa = 11.84DD236 pKa = 3.28GSMVV240 pKa = 3.03
Molecular weight: 25.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5SYC6|S5SYC6_9CIRC Replication-associated protein OS=Circoviridae 6 LDMD-2013 OX=1379710 PE=4 SV=1
MM1 pKa = 7.8PLRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84PVAMRR11 pKa = 11.84APRR14 pKa = 11.84PSLTAMRR21 pKa = 11.84ALGIPLRR28 pKa = 11.84AAPYY32 pKa = 8.21VARR35 pKa = 11.84AAGAVPGIAVAGLANLYY52 pKa = 9.79QSYY55 pKa = 9.65RR56 pKa = 11.84ATEE59 pKa = 3.77DD60 pKa = 2.85HH61 pKa = 6.64MARR64 pKa = 11.84TLKK67 pKa = 10.36RR68 pKa = 11.84GRR70 pKa = 11.84STYY73 pKa = 10.96GGGSSAKK80 pKa = 9.94KK81 pKa = 8.96YY82 pKa = 10.04KK83 pKa = 10.26KK84 pKa = 9.97PNGSRR89 pKa = 11.84WRR91 pKa = 11.84PGARR95 pKa = 11.84NWTSRR100 pKa = 11.84RR101 pKa = 11.84KK102 pKa = 9.52VAAKK106 pKa = 10.28LSAVEE111 pKa = 4.28TKK113 pKa = 10.39ISPLFRR119 pKa = 11.84RR120 pKa = 11.84EE121 pKa = 3.87DD122 pKa = 3.4VAATPFPTPDD132 pKa = 2.88TGNVVKK138 pKa = 9.96TGQDD142 pKa = 3.51LLTAAHH148 pKa = 6.25FVIGSGLPAAKK159 pKa = 9.79LQWYY163 pKa = 7.85TGVEE167 pKa = 4.21STATSITHH175 pKa = 6.07SLGGMFFPPAQHH187 pKa = 5.56TTGTDD192 pKa = 3.5EE193 pKa = 4.24VPSYY197 pKa = 10.7ADD199 pKa = 3.15RR200 pKa = 11.84MIRR203 pKa = 11.84NGRR206 pKa = 11.84SIYY209 pKa = 10.44LRR211 pKa = 11.84RR212 pKa = 11.84TSIAVHH218 pKa = 5.32ITCPTVQASGNLEE231 pKa = 4.15LNEE234 pKa = 4.41AGPFRR239 pKa = 11.84VRR241 pKa = 11.84MIVYY245 pKa = 9.49RR246 pKa = 11.84VRR248 pKa = 11.84LATTQSEE255 pKa = 4.38DD256 pKa = 3.37AEE258 pKa = 4.23EE259 pKa = 4.89RR260 pKa = 11.84DD261 pKa = 3.3TGKK264 pKa = 10.54DD265 pKa = 3.45FFIDD269 pKa = 3.41PMGRR273 pKa = 11.84PWGWKK278 pKa = 10.34GFDD281 pKa = 4.12DD282 pKa = 5.07QVKK285 pKa = 9.77QPDD288 pKa = 3.45THH290 pKa = 7.22LPRR293 pKa = 11.84ITGYY297 pKa = 11.01DD298 pKa = 3.61VMSQPVNSNKK308 pKa = 9.9YY309 pKa = 8.28QVIKK313 pKa = 10.46DD314 pKa = 3.87VKK316 pKa = 10.62FSMQPKK322 pKa = 8.43MSISAAEE329 pKa = 4.1SDD331 pKa = 3.76TTGQWLVNTPKK342 pKa = 10.55RR343 pKa = 11.84APHH346 pKa = 5.48TKK348 pKa = 9.95YY349 pKa = 10.87FNIQLPWGRR358 pKa = 11.84KK359 pKa = 6.6AHH361 pKa = 5.48FTNTITGAQEE371 pKa = 3.35DD372 pKa = 4.62TLALRR377 pKa = 11.84HH378 pKa = 5.63AAMMQEE384 pKa = 4.31SCDD387 pKa = 3.71FSPILKK393 pKa = 10.58SIDD396 pKa = 3.36LSAAEE401 pKa = 4.67PDD403 pKa = 3.56DD404 pKa = 4.61FNWRR408 pKa = 11.84THH410 pKa = 4.92IVFFGGPQRR419 pKa = 11.84GAGRR423 pKa = 11.84FTGTWEE429 pKa = 3.56PAITIRR435 pKa = 11.84GMTRR439 pKa = 11.84AYY441 pKa = 10.47DD442 pKa = 3.33AA443 pKa = 5.03
MM1 pKa = 7.8PLRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84PVAMRR11 pKa = 11.84APRR14 pKa = 11.84PSLTAMRR21 pKa = 11.84ALGIPLRR28 pKa = 11.84AAPYY32 pKa = 8.21VARR35 pKa = 11.84AAGAVPGIAVAGLANLYY52 pKa = 9.79QSYY55 pKa = 9.65RR56 pKa = 11.84ATEE59 pKa = 3.77DD60 pKa = 2.85HH61 pKa = 6.64MARR64 pKa = 11.84TLKK67 pKa = 10.36RR68 pKa = 11.84GRR70 pKa = 11.84STYY73 pKa = 10.96GGGSSAKK80 pKa = 9.94KK81 pKa = 8.96YY82 pKa = 10.04KK83 pKa = 10.26KK84 pKa = 9.97PNGSRR89 pKa = 11.84WRR91 pKa = 11.84PGARR95 pKa = 11.84NWTSRR100 pKa = 11.84RR101 pKa = 11.84KK102 pKa = 9.52VAAKK106 pKa = 10.28LSAVEE111 pKa = 4.28TKK113 pKa = 10.39ISPLFRR119 pKa = 11.84RR120 pKa = 11.84EE121 pKa = 3.87DD122 pKa = 3.4VAATPFPTPDD132 pKa = 2.88TGNVVKK138 pKa = 9.96TGQDD142 pKa = 3.51LLTAAHH148 pKa = 6.25FVIGSGLPAAKK159 pKa = 9.79LQWYY163 pKa = 7.85TGVEE167 pKa = 4.21STATSITHH175 pKa = 6.07SLGGMFFPPAQHH187 pKa = 5.56TTGTDD192 pKa = 3.5EE193 pKa = 4.24VPSYY197 pKa = 10.7ADD199 pKa = 3.15RR200 pKa = 11.84MIRR203 pKa = 11.84NGRR206 pKa = 11.84SIYY209 pKa = 10.44LRR211 pKa = 11.84RR212 pKa = 11.84TSIAVHH218 pKa = 5.32ITCPTVQASGNLEE231 pKa = 4.15LNEE234 pKa = 4.41AGPFRR239 pKa = 11.84VRR241 pKa = 11.84MIVYY245 pKa = 9.49RR246 pKa = 11.84VRR248 pKa = 11.84LATTQSEE255 pKa = 4.38DD256 pKa = 3.37AEE258 pKa = 4.23EE259 pKa = 4.89RR260 pKa = 11.84DD261 pKa = 3.3TGKK264 pKa = 10.54DD265 pKa = 3.45FFIDD269 pKa = 3.41PMGRR273 pKa = 11.84PWGWKK278 pKa = 10.34GFDD281 pKa = 4.12DD282 pKa = 5.07QVKK285 pKa = 9.77QPDD288 pKa = 3.45THH290 pKa = 7.22LPRR293 pKa = 11.84ITGYY297 pKa = 11.01DD298 pKa = 3.61VMSQPVNSNKK308 pKa = 9.9YY309 pKa = 8.28QVIKK313 pKa = 10.46DD314 pKa = 3.87VKK316 pKa = 10.62FSMQPKK322 pKa = 8.43MSISAAEE329 pKa = 4.1SDD331 pKa = 3.76TTGQWLVNTPKK342 pKa = 10.55RR343 pKa = 11.84APHH346 pKa = 5.48TKK348 pKa = 9.95YY349 pKa = 10.87FNIQLPWGRR358 pKa = 11.84KK359 pKa = 6.6AHH361 pKa = 5.48FTNTITGAQEE371 pKa = 3.35DD372 pKa = 4.62TLALRR377 pKa = 11.84HH378 pKa = 5.63AAMMQEE384 pKa = 4.31SCDD387 pKa = 3.71FSPILKK393 pKa = 10.58SIDD396 pKa = 3.36LSAAEE401 pKa = 4.67PDD403 pKa = 3.56DD404 pKa = 4.61FNWRR408 pKa = 11.84THH410 pKa = 4.92IVFFGGPQRR419 pKa = 11.84GAGRR423 pKa = 11.84FTGTWEE429 pKa = 3.56PAITIRR435 pKa = 11.84GMTRR439 pKa = 11.84AYY441 pKa = 10.47DD442 pKa = 3.33AA443 pKa = 5.03
Molecular weight: 49.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
862 |
179 |
443 |
287.3 |
31.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.397 ± 1.306 | 1.16 ± 0.565 |
4.524 ± 0.314 | 4.408 ± 0.796 |
3.712 ± 0.327 | 8.817 ± 1.093 |
2.784 ± 0.71 | 3.712 ± 0.76 |
4.06 ± 0.617 | 6.729 ± 1.409 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.784 ± 0.298 | 2.204 ± 0.516 |
7.541 ± 0.84 | 4.06 ± 0.362 |
8.237 ± 0.401 | 6.497 ± 0.123 |
7.425 ± 1.11 | 6.497 ± 0.758 |
2.32 ± 0.503 | 3.132 ± 0.138 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
