
Torque teno Leptonychotes weddellii virus-1
Taxonomy: Viruses; Anelloviridae; Lambdatorquevirus; Torque teno pinniped virus 8
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
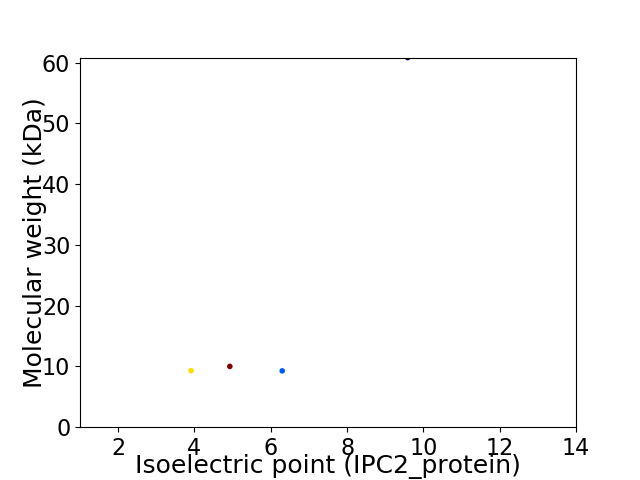
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2RVW0|A0A1Z2RVW0_9VIRU ORF2 OS=Torque teno Leptonychotes weddellii virus-1 OX=2012676 PE=4 SV=1
MM1 pKa = 7.24ISYY4 pKa = 10.5RR5 pKa = 11.84EE6 pKa = 3.93TSTQTEE12 pKa = 4.5CSRR15 pKa = 11.84KK16 pKa = 9.19EE17 pKa = 3.85LMRR20 pKa = 11.84EE21 pKa = 3.6LLEE24 pKa = 5.55IIDD27 pKa = 3.93VTSQPEE33 pKa = 3.94WGMSPIGSPPEE44 pKa = 4.15SEE46 pKa = 4.35SDD48 pKa = 3.3SDD50 pKa = 4.4SLIEE54 pKa = 4.12NSSTGSGSSSEE65 pKa = 4.96FYY67 pKa = 10.46WEE69 pKa = 5.41DD70 pKa = 3.13EE71 pKa = 4.4TNPGGEE77 pKa = 4.56GPPLVPPQQ85 pKa = 3.45
MM1 pKa = 7.24ISYY4 pKa = 10.5RR5 pKa = 11.84EE6 pKa = 3.93TSTQTEE12 pKa = 4.5CSRR15 pKa = 11.84KK16 pKa = 9.19EE17 pKa = 3.85LMRR20 pKa = 11.84EE21 pKa = 3.6LLEE24 pKa = 5.55IIDD27 pKa = 3.93VTSQPEE33 pKa = 3.94WGMSPIGSPPEE44 pKa = 4.15SEE46 pKa = 4.35SDD48 pKa = 3.3SDD50 pKa = 4.4SLIEE54 pKa = 4.12NSSTGSGSSSEE65 pKa = 4.96FYY67 pKa = 10.46WEE69 pKa = 5.41DD70 pKa = 3.13EE71 pKa = 4.4TNPGGEE77 pKa = 4.56GPPLVPPQQ85 pKa = 3.45
Molecular weight: 9.31 kDa
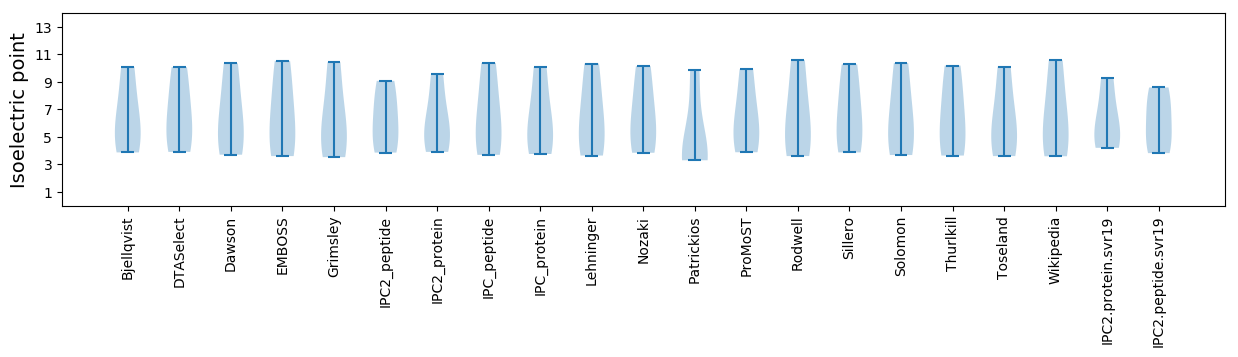
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2RVY6|A0A1Z2RVY6_9VIRU Capsid protein OS=Torque teno Leptonychotes weddellii virus-1 OX=2012676 PE=3 SV=1
MM1 pKa = 7.84AYY3 pKa = 10.03RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84WRR9 pKa = 11.84KK10 pKa = 7.57PRR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84WRR17 pKa = 11.84TYY19 pKa = 9.7RR20 pKa = 11.84RR21 pKa = 11.84NRR23 pKa = 11.84YY24 pKa = 5.39WRR26 pKa = 11.84RR27 pKa = 11.84PHH29 pKa = 5.97RR30 pKa = 11.84RR31 pKa = 11.84HH32 pKa = 4.26WRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 9.27IRR40 pKa = 11.84PRR42 pKa = 11.84TATVRR47 pKa = 11.84YY48 pKa = 8.07YY49 pKa = 10.09PSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84KK55 pKa = 9.59RR56 pKa = 11.84ISVRR60 pKa = 11.84GWEE63 pKa = 4.02PLGNVCSSDD72 pKa = 3.19IASSEE77 pKa = 3.88ATPYY81 pKa = 10.53TDD83 pKa = 5.02LDD85 pKa = 3.97MYY87 pKa = 11.17DD88 pKa = 4.73LSPLEE93 pKa = 4.78DD94 pKa = 3.42DD95 pKa = 3.27TRR97 pKa = 11.84KK98 pKa = 10.65YY99 pKa = 9.38KK100 pKa = 10.89GKK102 pKa = 8.35WHH104 pKa = 5.96GTWGHH109 pKa = 6.25HH110 pKa = 5.49FFTLRR115 pKa = 11.84SLLVRR120 pKa = 11.84AKK122 pKa = 10.79YY123 pKa = 9.84YY124 pKa = 10.56FNYY127 pKa = 9.34WSSDD131 pKa = 2.91WEE133 pKa = 4.56GYY135 pKa = 10.88DD136 pKa = 3.4YY137 pKa = 11.83VKK139 pKa = 10.96FKK141 pKa = 10.99GGYY144 pKa = 7.56IWIPRR149 pKa = 11.84MTYY152 pKa = 10.13FSWIFYY158 pKa = 9.97LDD160 pKa = 3.53SSIQSNPRR168 pKa = 11.84EE169 pKa = 4.34DD170 pKa = 4.3GEE172 pKa = 4.33PEE174 pKa = 3.7AKK176 pKa = 10.05YY177 pKa = 10.28KK178 pKa = 10.55YY179 pKa = 8.95EE180 pKa = 4.28KK181 pKa = 10.3SWLHH185 pKa = 6.31PGILLNRR192 pKa = 11.84PGSKK196 pKa = 10.56LMISSLQQPNRR207 pKa = 11.84PFYY210 pKa = 10.8RR211 pKa = 11.84RR212 pKa = 11.84IKK214 pKa = 8.54VTPPSAWEE222 pKa = 3.75GNYY225 pKa = 10.57RR226 pKa = 11.84MDD228 pKa = 3.3TAMDD232 pKa = 3.61YY233 pKa = 11.58LLFHH237 pKa = 7.05WGWTVCNVTASFFDD251 pKa = 4.35FFCQRR256 pKa = 11.84KK257 pKa = 9.28RR258 pKa = 11.84SATHH262 pKa = 6.76NDD264 pKa = 3.39TCPTTPWFIHH274 pKa = 6.07DD275 pKa = 4.27SCWKK279 pKa = 10.03QGITANEE286 pKa = 3.73VCKK289 pKa = 10.45AIKK292 pKa = 9.6EE293 pKa = 4.28VYY295 pKa = 10.02DD296 pKa = 4.67RR297 pKa = 11.84INKK300 pKa = 8.93RR301 pKa = 11.84HH302 pKa = 6.56LDD304 pKa = 3.66PRR306 pKa = 11.84SVWVNRR312 pKa = 11.84KK313 pKa = 8.34MYY315 pKa = 10.41IKK317 pKa = 9.84SDD319 pKa = 3.76CVNTPNSNSVPSNCHH334 pKa = 3.44NWGPFLPQNVILEE347 pKa = 4.45TSTGNSLYY355 pKa = 10.33FRR357 pKa = 11.84YY358 pKa = 10.29KK359 pKa = 10.73LFFEE363 pKa = 4.55VSGDD367 pKa = 3.24SLYY370 pKa = 10.72RR371 pKa = 11.84RR372 pKa = 11.84QPSQPCKK379 pKa = 10.74DD380 pKa = 3.58GVIPPAPGAEE390 pKa = 4.33SACPEE395 pKa = 3.47ISTRR399 pKa = 11.84SIYY402 pKa = 10.17KK403 pKa = 9.77KK404 pKa = 10.14KK405 pKa = 10.3KK406 pKa = 8.92RR407 pKa = 11.84PQSIYY412 pKa = 10.75DD413 pKa = 3.62ILQGDD418 pKa = 4.61LDD420 pKa = 4.41SDD422 pKa = 3.81GMLTEE427 pKa = 4.41RR428 pKa = 11.84AYY430 pKa = 10.5EE431 pKa = 4.52RR432 pKa = 11.84ITGDD436 pKa = 3.45NRR438 pKa = 11.84CDD440 pKa = 3.23QPTRR444 pKa = 11.84MGDD447 pKa = 3.35VPHH450 pKa = 7.05RR451 pKa = 11.84VPPRR455 pKa = 11.84KK456 pKa = 9.1RR457 pKa = 11.84VRR459 pKa = 11.84FRR461 pKa = 11.84LSDD464 pKa = 3.28RR465 pKa = 11.84KK466 pKa = 9.39LQHH469 pKa = 6.8RR470 pKa = 11.84KK471 pKa = 9.72RR472 pKa = 11.84EE473 pKa = 4.28LIRR476 pKa = 11.84ILLGRR481 pKa = 11.84RR482 pKa = 11.84DD483 pKa = 3.57QPRR486 pKa = 11.84GGGSPPSPPPVEE498 pKa = 4.35EE499 pKa = 4.6PLDD502 pKa = 3.86LLLNFPKK509 pKa = 10.71
MM1 pKa = 7.84AYY3 pKa = 10.03RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84WRR9 pKa = 11.84KK10 pKa = 7.57PRR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84WRR17 pKa = 11.84TYY19 pKa = 9.7RR20 pKa = 11.84RR21 pKa = 11.84NRR23 pKa = 11.84YY24 pKa = 5.39WRR26 pKa = 11.84RR27 pKa = 11.84PHH29 pKa = 5.97RR30 pKa = 11.84RR31 pKa = 11.84HH32 pKa = 4.26WRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 9.27IRR40 pKa = 11.84PRR42 pKa = 11.84TATVRR47 pKa = 11.84YY48 pKa = 8.07YY49 pKa = 10.09PSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84KK55 pKa = 9.59RR56 pKa = 11.84ISVRR60 pKa = 11.84GWEE63 pKa = 4.02PLGNVCSSDD72 pKa = 3.19IASSEE77 pKa = 3.88ATPYY81 pKa = 10.53TDD83 pKa = 5.02LDD85 pKa = 3.97MYY87 pKa = 11.17DD88 pKa = 4.73LSPLEE93 pKa = 4.78DD94 pKa = 3.42DD95 pKa = 3.27TRR97 pKa = 11.84KK98 pKa = 10.65YY99 pKa = 9.38KK100 pKa = 10.89GKK102 pKa = 8.35WHH104 pKa = 5.96GTWGHH109 pKa = 6.25HH110 pKa = 5.49FFTLRR115 pKa = 11.84SLLVRR120 pKa = 11.84AKK122 pKa = 10.79YY123 pKa = 9.84YY124 pKa = 10.56FNYY127 pKa = 9.34WSSDD131 pKa = 2.91WEE133 pKa = 4.56GYY135 pKa = 10.88DD136 pKa = 3.4YY137 pKa = 11.83VKK139 pKa = 10.96FKK141 pKa = 10.99GGYY144 pKa = 7.56IWIPRR149 pKa = 11.84MTYY152 pKa = 10.13FSWIFYY158 pKa = 9.97LDD160 pKa = 3.53SSIQSNPRR168 pKa = 11.84EE169 pKa = 4.34DD170 pKa = 4.3GEE172 pKa = 4.33PEE174 pKa = 3.7AKK176 pKa = 10.05YY177 pKa = 10.28KK178 pKa = 10.55YY179 pKa = 8.95EE180 pKa = 4.28KK181 pKa = 10.3SWLHH185 pKa = 6.31PGILLNRR192 pKa = 11.84PGSKK196 pKa = 10.56LMISSLQQPNRR207 pKa = 11.84PFYY210 pKa = 10.8RR211 pKa = 11.84RR212 pKa = 11.84IKK214 pKa = 8.54VTPPSAWEE222 pKa = 3.75GNYY225 pKa = 10.57RR226 pKa = 11.84MDD228 pKa = 3.3TAMDD232 pKa = 3.61YY233 pKa = 11.58LLFHH237 pKa = 7.05WGWTVCNVTASFFDD251 pKa = 4.35FFCQRR256 pKa = 11.84KK257 pKa = 9.28RR258 pKa = 11.84SATHH262 pKa = 6.76NDD264 pKa = 3.39TCPTTPWFIHH274 pKa = 6.07DD275 pKa = 4.27SCWKK279 pKa = 10.03QGITANEE286 pKa = 3.73VCKK289 pKa = 10.45AIKK292 pKa = 9.6EE293 pKa = 4.28VYY295 pKa = 10.02DD296 pKa = 4.67RR297 pKa = 11.84INKK300 pKa = 8.93RR301 pKa = 11.84HH302 pKa = 6.56LDD304 pKa = 3.66PRR306 pKa = 11.84SVWVNRR312 pKa = 11.84KK313 pKa = 8.34MYY315 pKa = 10.41IKK317 pKa = 9.84SDD319 pKa = 3.76CVNTPNSNSVPSNCHH334 pKa = 3.44NWGPFLPQNVILEE347 pKa = 4.45TSTGNSLYY355 pKa = 10.33FRR357 pKa = 11.84YY358 pKa = 10.29KK359 pKa = 10.73LFFEE363 pKa = 4.55VSGDD367 pKa = 3.24SLYY370 pKa = 10.72RR371 pKa = 11.84RR372 pKa = 11.84QPSQPCKK379 pKa = 10.74DD380 pKa = 3.58GVIPPAPGAEE390 pKa = 4.33SACPEE395 pKa = 3.47ISTRR399 pKa = 11.84SIYY402 pKa = 10.17KK403 pKa = 9.77KK404 pKa = 10.14KK405 pKa = 10.3KK406 pKa = 8.92RR407 pKa = 11.84PQSIYY412 pKa = 10.75DD413 pKa = 3.62ILQGDD418 pKa = 4.61LDD420 pKa = 4.41SDD422 pKa = 3.81GMLTEE427 pKa = 4.41RR428 pKa = 11.84AYY430 pKa = 10.5EE431 pKa = 4.52RR432 pKa = 11.84ITGDD436 pKa = 3.45NRR438 pKa = 11.84CDD440 pKa = 3.23QPTRR444 pKa = 11.84MGDD447 pKa = 3.35VPHH450 pKa = 7.05RR451 pKa = 11.84VPPRR455 pKa = 11.84KK456 pKa = 9.1RR457 pKa = 11.84VRR459 pKa = 11.84FRR461 pKa = 11.84LSDD464 pKa = 3.28RR465 pKa = 11.84KK466 pKa = 9.39LQHH469 pKa = 6.8RR470 pKa = 11.84KK471 pKa = 9.72RR472 pKa = 11.84EE473 pKa = 4.28LIRR476 pKa = 11.84ILLGRR481 pKa = 11.84RR482 pKa = 11.84DD483 pKa = 3.57QPRR486 pKa = 11.84GGGSPPSPPPVEE498 pKa = 4.35EE499 pKa = 4.6PLDD502 pKa = 3.86LLLNFPKK509 pKa = 10.71
Molecular weight: 60.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
767 |
80 |
509 |
191.8 |
22.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.738 ± 0.961 | 2.347 ± 0.44 |
5.737 ± 0.572 | 5.606 ± 2.174 |
3.781 ± 0.843 | 7.301 ± 2.728 |
2.608 ± 0.642 | 5.215 ± 0.845 |
4.954 ± 1.079 | 7.823 ± 2.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.956 ± 0.382 | 3.651 ± 0.533 |
7.953 ± 0.876 | 2.999 ± 0.635 |
9.778 ± 3.304 | 8.735 ± 2.099 |
4.694 ± 0.824 | 4.042 ± 1.086 |
3.259 ± 0.525 | 4.824 ± 1.186 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
