
Cecembia lonarensis LW9
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Cecembia; Cecembia lonarensis
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
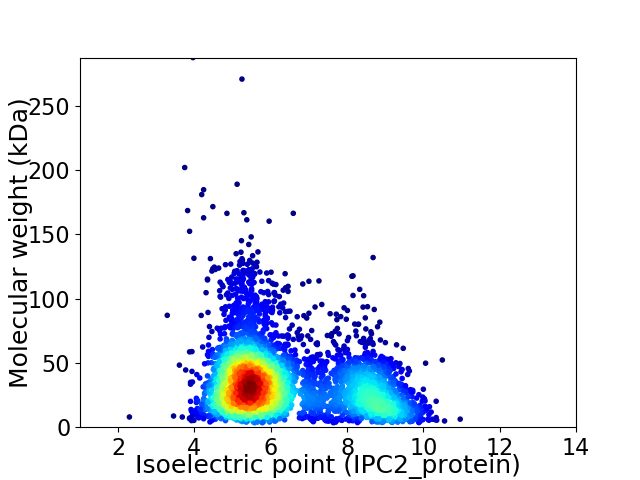
Virtual 2D-PAGE plot for 4213 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K1L266|K1L266_9BACT Macrolide export ATP-binding/permease protein MacB OS=Cecembia lonarensis LW9 OX=1225176 GN=macB_1 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 10.21RR3 pKa = 11.84YY4 pKa = 9.65IKK6 pKa = 9.25YY7 pKa = 9.08TNINLFFIALGLLIIFGNHH26 pKa = 4.11QKK28 pKa = 10.61VYY30 pKa = 10.97AQGFNNNQWIFGYY43 pKa = 10.29CGSGQEE49 pKa = 3.96NSYY52 pKa = 11.3LSFGRR57 pKa = 11.84GEE59 pKa = 4.27DD60 pKa = 3.56PVIRR64 pKa = 11.84TLPPSILIGDD74 pKa = 3.82EE75 pKa = 3.95NGNVDD80 pKa = 3.49NNAIVIDD87 pKa = 5.1PITGQILFYY96 pKa = 10.41TNGVLVYY103 pKa = 10.4NYY105 pKa = 9.86DD106 pKa = 3.53DD107 pKa = 5.0QIIQGAPNGINGQTDD122 pKa = 3.01VRR124 pKa = 11.84QTVAIGTLNYY134 pKa = 9.49EE135 pKa = 4.28AEE137 pKa = 4.59GDD139 pKa = 3.58RR140 pKa = 11.84QFYY143 pKa = 10.19IFHH146 pKa = 6.59ITPGGQLLYY155 pKa = 10.86SVVDD159 pKa = 3.71MNSPGGAQGNAPPLGQVIAFDD180 pKa = 3.68QLLSNNVSGAISVVKK195 pKa = 9.67TPQSPSYY202 pKa = 10.34LISFEE207 pKa = 3.94GGEE210 pKa = 4.06LVARR214 pKa = 11.84RR215 pKa = 11.84VEE217 pKa = 4.15ANQGDD222 pKa = 4.25FTITDD227 pKa = 3.27NFGFPFTPKK236 pKa = 10.52AIIFDD241 pKa = 3.85EE242 pKa = 4.28SGKK245 pKa = 10.77LILIPEE251 pKa = 4.52GDD253 pKa = 3.95GEE255 pKa = 4.97DD256 pKa = 3.47IAVLDD261 pKa = 4.62FDD263 pKa = 4.39TSTGSFGNLSFISNSGGNAAIEE285 pKa = 4.29GAAFSPDD292 pKa = 2.56GDD294 pKa = 3.9FIYY297 pKa = 10.27FSRR300 pKa = 11.84GDD302 pKa = 3.27QLFRR306 pKa = 11.84VPANDD311 pKa = 4.81LDD313 pKa = 4.1TDD315 pKa = 4.07PEE317 pKa = 4.46VIPLEE322 pKa = 4.12GDD324 pKa = 2.4IDD326 pKa = 5.12AIFDD330 pKa = 3.68IKK332 pKa = 10.94VGPDD336 pKa = 2.69GRR338 pKa = 11.84LYY340 pKa = 10.73YY341 pKa = 9.88IYY343 pKa = 10.13EE344 pKa = 4.14EE345 pKa = 4.7TPGGPQLIGRR355 pKa = 11.84VNNPDD360 pKa = 3.6EE361 pKa = 4.59EE362 pKa = 4.42DD363 pKa = 3.93LEE365 pKa = 4.18EE366 pKa = 6.04LDD368 pKa = 4.98IEE370 pKa = 4.19EE371 pKa = 5.3DD372 pKa = 3.74PFNGTDD378 pKa = 3.4FCGRR382 pKa = 11.84VFPQFAPNQDD392 pKa = 3.1INPTVDD398 pKa = 5.18FDD400 pKa = 3.97WEE402 pKa = 4.27PDD404 pKa = 4.16FPCSNNPVQLTSQITPEE421 pKa = 3.8NYY423 pKa = 9.55RR424 pKa = 11.84PVSFEE429 pKa = 3.71WTFEE433 pKa = 4.05PPLTDD438 pKa = 3.35EE439 pKa = 5.27DD440 pKa = 4.69GEE442 pKa = 4.3PVDD445 pKa = 4.08IDD447 pKa = 3.81FNQEE451 pKa = 3.34HH452 pKa = 6.79LLIPEE457 pKa = 4.33EE458 pKa = 4.06ATANQSIQVTLTVTFADD475 pKa = 4.23GNVVTVPKK483 pKa = 9.91TIQLTEE489 pKa = 4.06NNLEE493 pKa = 4.42ANFSAQDD500 pKa = 3.42TTICEE505 pKa = 4.7GPCVDD510 pKa = 5.04IGSLLQVQEE519 pKa = 4.41GGGQGGEE526 pKa = 4.12NPNVGGPGQGGNYY539 pKa = 9.85EE540 pKa = 4.36YY541 pKa = 10.62FWSNRR546 pKa = 11.84RR547 pKa = 11.84DD548 pKa = 3.74EE549 pKa = 4.42GWVRR553 pKa = 11.84DD554 pKa = 3.56TDD556 pKa = 3.76NCVDD560 pKa = 3.66RR561 pKa = 11.84PGLYY565 pKa = 8.66WVLVRR570 pKa = 11.84EE571 pKa = 4.7PGSEE575 pKa = 3.97CYY577 pKa = 10.14AYY579 pKa = 10.63AEE581 pKa = 4.08VRR583 pKa = 11.84VRR585 pKa = 11.84IWDD588 pKa = 4.75LPDD591 pKa = 3.0QSNNVWLFGNGAGLDD606 pKa = 4.04FNPDD610 pKa = 3.58PDD612 pKa = 5.11DD613 pKa = 4.22EE614 pKa = 4.43EE615 pKa = 6.48APLPRR620 pKa = 11.84PVSHH624 pKa = 6.64NRR626 pKa = 11.84DD627 pKa = 3.09IPAGTTTISDD637 pKa = 3.26EE638 pKa = 4.24TGQVLFFTDD647 pKa = 5.18GEE649 pKa = 4.95SVWDD653 pKa = 4.04LNGDD657 pKa = 4.3LMANGDD663 pKa = 4.1NIGGNNQASEE673 pKa = 4.39GVIAVPVPQNQTIFYY688 pKa = 10.36VFTTQRR694 pKa = 11.84AADD697 pKa = 3.66GSNRR701 pKa = 11.84VSYY704 pKa = 10.91SVVDD708 pKa = 3.76IKK710 pKa = 10.38TDD712 pKa = 3.08NPTGVGSVVTKK723 pKa = 11.13NNFLFSPSTEE733 pKa = 3.72QTAALASGDD742 pKa = 3.95TTWVLFHH749 pKa = 7.06EE750 pKa = 5.11LGNNTFRR757 pKa = 11.84AYY759 pKa = 9.44PVSPFGIGSPVFSSVGSNHH778 pKa = 6.05NFTSGVGSMKK788 pKa = 10.45FSPDD792 pKa = 3.02GNQVAVTISDD802 pKa = 4.02GNCSRR807 pKa = 11.84LEE809 pKa = 3.56IFDD812 pKa = 4.87FNEE815 pKa = 3.57QTGRR819 pKa = 11.84LTEE822 pKa = 3.88YY823 pKa = 11.39ALIEE827 pKa = 4.34LGCGAEE833 pKa = 3.96EE834 pKa = 4.76VYY836 pKa = 10.94GLEE839 pKa = 4.09FSPDD843 pKa = 2.86GSRR846 pKa = 11.84IFVSYY851 pKa = 9.58TGSPGKK857 pKa = 9.98IEE859 pKa = 4.27EE860 pKa = 4.39YY861 pKa = 9.99LVKK864 pKa = 10.48RR865 pKa = 11.84PEE867 pKa = 4.1TAVEE871 pKa = 4.32GEE873 pKa = 5.19DD874 pKa = 3.28IDD876 pKa = 5.83SDD878 pKa = 4.48LSCFDD883 pKa = 3.85CFEE886 pKa = 4.34NASNRR891 pKa = 11.84SQRR894 pKa = 11.84EE895 pKa = 3.38NCIIDD900 pKa = 3.58SRR902 pKa = 11.84NVLSTAGPFGAIQIGPDD919 pKa = 3.28GQIYY923 pKa = 8.61VARR926 pKa = 11.84PDD928 pKa = 3.62RR929 pKa = 11.84NEE931 pKa = 3.36ITAILPGFNCLDD943 pKa = 3.25SFYY946 pKa = 10.96NEE948 pKa = 4.08QGVSLLPGTSMNLGLPAFVQQSGSSIPEE976 pKa = 3.67PALAGIEE983 pKa = 4.07RR984 pKa = 11.84LCFDD988 pKa = 4.43PDD990 pKa = 3.27GTIGLFEE997 pKa = 5.74AGGEE1001 pKa = 4.0PDD1003 pKa = 2.99IDD1005 pKa = 4.01SYY1007 pKa = 11.62FWTITHH1013 pKa = 6.97EE1014 pKa = 4.53DD1015 pKa = 3.91GEE1017 pKa = 5.16VVFEE1021 pKa = 4.24EE1022 pKa = 5.04GGPGEE1027 pKa = 4.34TFQEE1031 pKa = 4.24IEE1033 pKa = 4.02PLFPRR1038 pKa = 11.84VGIYY1042 pKa = 8.97TVALRR1047 pKa = 11.84VDD1049 pKa = 3.21RR1050 pKa = 11.84CGNPEE1055 pKa = 3.89YY1056 pKa = 10.37YY1057 pKa = 9.6RR1058 pKa = 11.84EE1059 pKa = 4.11SMEE1062 pKa = 4.19VEE1064 pKa = 4.64VIASPEE1070 pKa = 3.82LTLPEE1075 pKa = 4.35EE1076 pKa = 4.26ATLCVGTPVPLTAIDD1091 pKa = 4.93GYY1093 pKa = 11.26DD1094 pKa = 3.69PSEE1097 pKa = 3.87GLYY1100 pKa = 10.91DD1101 pKa = 3.92FEE1103 pKa = 4.48WRR1105 pKa = 11.84NAAGQLIGDD1114 pKa = 4.19EE1115 pKa = 4.15NSNTIEE1121 pKa = 4.03VVEE1124 pKa = 4.17EE1125 pKa = 3.83SVYY1128 pKa = 10.17TVTVRR1133 pKa = 11.84FRR1135 pKa = 11.84VPEE1138 pKa = 4.19GVDD1141 pKa = 3.2PEE1143 pKa = 4.78EE1144 pKa = 5.01FDD1146 pKa = 3.52ACPVSRR1152 pKa = 11.84SIFVGPAFDD1161 pKa = 4.66FDD1163 pKa = 5.56LIQSAEE1169 pKa = 4.3EE1170 pKa = 4.12VCLDD1174 pKa = 3.63EE1175 pKa = 4.6PVVIFSPNTPITGEE1189 pKa = 3.9WFFRR1193 pKa = 11.84PAGSPDD1199 pKa = 3.32RR1200 pKa = 11.84VSLGEE1205 pKa = 3.94FFEE1208 pKa = 5.64LEE1210 pKa = 4.9LIPASLPSPGDD1221 pKa = 3.3YY1222 pKa = 10.73EE1223 pKa = 4.79IIFFAEE1229 pKa = 4.27DD1230 pKa = 3.9PLVEE1234 pKa = 4.22GCFVEE1239 pKa = 5.65KK1240 pKa = 10.28IVPLTIFPLPAVDD1253 pKa = 5.73AVILSDD1259 pKa = 5.71AEE1261 pKa = 4.11DD1262 pKa = 4.29CEE1264 pKa = 4.73IPDD1267 pKa = 3.61GSFEE1271 pKa = 3.98ITALSDD1277 pKa = 3.45IEE1279 pKa = 4.19SLEE1282 pKa = 3.88VLEE1285 pKa = 5.39LGLLFGPIAAGEE1297 pKa = 4.07ALPPFTDD1304 pKa = 4.8LEE1306 pKa = 4.77PGLYY1310 pKa = 8.48TLQMLNGFGCEE1321 pKa = 4.77FIQSVTIRR1329 pKa = 11.84NLNPPRR1335 pKa = 11.84GIEE1338 pKa = 4.26GYY1340 pKa = 8.36EE1341 pKa = 3.87VEE1343 pKa = 4.92VSPEE1347 pKa = 3.63ICDD1350 pKa = 3.49VNGVLDD1356 pKa = 4.07GQLTIRR1362 pKa = 11.84FISGGASGTYY1372 pKa = 9.93QITRR1376 pKa = 11.84QGDD1379 pKa = 3.61GAFVSDD1385 pKa = 3.77NFNDD1389 pKa = 3.59VEE1391 pKa = 4.63EE1392 pKa = 4.34ITVPVPAGIYY1402 pKa = 9.39SVVVEE1407 pKa = 4.96DD1408 pKa = 4.16VDD1410 pKa = 4.07GCAVPFEE1417 pKa = 4.71DD1418 pKa = 3.78LVEE1421 pKa = 4.15VEE1423 pKa = 4.33EE1424 pKa = 4.35VEE1426 pKa = 4.37IVAFSAPGNVEE1437 pKa = 3.61ACEE1440 pKa = 4.29SFTFSPQSEE1449 pKa = 4.55DD1450 pKa = 3.25EE1451 pKa = 4.21LVYY1454 pKa = 9.84TLRR1457 pKa = 11.84DD1458 pKa = 3.42AQGNQIQATPEE1469 pKa = 3.89GSFTITEE1476 pKa = 4.04AGLYY1480 pKa = 9.91VLTGEE1485 pKa = 4.9DD1486 pKa = 3.8PDD1488 pKa = 4.8GIKK1491 pKa = 10.34CARR1494 pKa = 11.84EE1495 pKa = 3.54RR1496 pKa = 11.84EE1497 pKa = 4.35INVTLTTAPVFSLAGPRR1514 pKa = 11.84VDD1516 pKa = 4.57CDD1518 pKa = 3.3LGLFYY1523 pKa = 10.72EE1524 pKa = 4.97AVLGPNEE1531 pKa = 4.94DD1532 pKa = 3.92PNQVFIFWLDD1542 pKa = 3.45SNGEE1546 pKa = 4.33VVSRR1550 pKa = 11.84NPLFFPRR1557 pKa = 11.84VPGTYY1562 pKa = 9.67FLEE1565 pKa = 4.34VQPRR1569 pKa = 11.84TGSLCEE1575 pKa = 3.84IDD1577 pKa = 4.85PISFTVEE1584 pKa = 3.86DD1585 pKa = 4.62TDD1587 pKa = 3.8PQIDD1591 pKa = 3.97VEE1593 pKa = 4.88LEE1595 pKa = 3.81ALPFCAEE1602 pKa = 4.54DD1603 pKa = 3.94PFTLISVIADD1613 pKa = 3.84LSDD1616 pKa = 3.15VALIQWFEE1624 pKa = 4.17IINGEE1629 pKa = 4.29RR1630 pKa = 11.84EE1631 pKa = 4.64LIEE1634 pKa = 4.52DD1635 pKa = 4.85WIGLDD1640 pKa = 3.16QVVALFEE1647 pKa = 4.54GVFEE1651 pKa = 4.53IVLINNDD1658 pKa = 2.85GCEE1661 pKa = 3.76IGTAIIEE1668 pKa = 4.22IRR1670 pKa = 11.84QSTIGPPDD1678 pKa = 3.98LDD1680 pKa = 3.75EE1681 pKa = 6.07LYY1683 pKa = 10.79VICAPEE1689 pKa = 4.99GILVEE1694 pKa = 4.56IGPGEE1699 pKa = 3.95YY1700 pKa = 10.42DD1701 pKa = 3.11NYY1703 pKa = 10.86QWILDD1708 pKa = 4.02EE1709 pKa = 4.33EE1710 pKa = 4.9VVAVSPTFSPTIPGDD1725 pKa = 3.5YY1726 pKa = 10.48EE1727 pKa = 5.84LIVSDD1732 pKa = 5.62DD1733 pKa = 3.94LGCEE1737 pKa = 3.7FRR1739 pKa = 11.84ARR1741 pKa = 11.84FVVEE1745 pKa = 4.29EE1746 pKa = 4.09DD1747 pKa = 4.04CEE1749 pKa = 4.44LKK1751 pKa = 10.79INFPNAIIPNDD1762 pKa = 3.49SEE1764 pKa = 5.02RR1765 pKa = 11.84NFVVYY1770 pKa = 10.49TNDD1773 pKa = 3.31FVDD1776 pKa = 3.52EE1777 pKa = 4.74LEE1779 pKa = 4.17VFIYY1783 pKa = 10.45NRR1785 pKa = 11.84WGEE1788 pKa = 4.22LIFYY1792 pKa = 10.26CEE1794 pKa = 4.29SNGVNGEE1801 pKa = 4.05IPRR1804 pKa = 11.84CAWDD1808 pKa = 4.37GIVNGRR1814 pKa = 11.84TVPIGTYY1821 pKa = 9.46PVVVNYY1827 pKa = 10.51GSTSQNVRR1835 pKa = 11.84KK1836 pKa = 7.68TLKK1839 pKa = 9.52RR1840 pKa = 11.84TILVIEE1846 pKa = 4.2
MM1 pKa = 7.33KK2 pKa = 10.21RR3 pKa = 11.84YY4 pKa = 9.65IKK6 pKa = 9.25YY7 pKa = 9.08TNINLFFIALGLLIIFGNHH26 pKa = 4.11QKK28 pKa = 10.61VYY30 pKa = 10.97AQGFNNNQWIFGYY43 pKa = 10.29CGSGQEE49 pKa = 3.96NSYY52 pKa = 11.3LSFGRR57 pKa = 11.84GEE59 pKa = 4.27DD60 pKa = 3.56PVIRR64 pKa = 11.84TLPPSILIGDD74 pKa = 3.82EE75 pKa = 3.95NGNVDD80 pKa = 3.49NNAIVIDD87 pKa = 5.1PITGQILFYY96 pKa = 10.41TNGVLVYY103 pKa = 10.4NYY105 pKa = 9.86DD106 pKa = 3.53DD107 pKa = 5.0QIIQGAPNGINGQTDD122 pKa = 3.01VRR124 pKa = 11.84QTVAIGTLNYY134 pKa = 9.49EE135 pKa = 4.28AEE137 pKa = 4.59GDD139 pKa = 3.58RR140 pKa = 11.84QFYY143 pKa = 10.19IFHH146 pKa = 6.59ITPGGQLLYY155 pKa = 10.86SVVDD159 pKa = 3.71MNSPGGAQGNAPPLGQVIAFDD180 pKa = 3.68QLLSNNVSGAISVVKK195 pKa = 9.67TPQSPSYY202 pKa = 10.34LISFEE207 pKa = 3.94GGEE210 pKa = 4.06LVARR214 pKa = 11.84RR215 pKa = 11.84VEE217 pKa = 4.15ANQGDD222 pKa = 4.25FTITDD227 pKa = 3.27NFGFPFTPKK236 pKa = 10.52AIIFDD241 pKa = 3.85EE242 pKa = 4.28SGKK245 pKa = 10.77LILIPEE251 pKa = 4.52GDD253 pKa = 3.95GEE255 pKa = 4.97DD256 pKa = 3.47IAVLDD261 pKa = 4.62FDD263 pKa = 4.39TSTGSFGNLSFISNSGGNAAIEE285 pKa = 4.29GAAFSPDD292 pKa = 2.56GDD294 pKa = 3.9FIYY297 pKa = 10.27FSRR300 pKa = 11.84GDD302 pKa = 3.27QLFRR306 pKa = 11.84VPANDD311 pKa = 4.81LDD313 pKa = 4.1TDD315 pKa = 4.07PEE317 pKa = 4.46VIPLEE322 pKa = 4.12GDD324 pKa = 2.4IDD326 pKa = 5.12AIFDD330 pKa = 3.68IKK332 pKa = 10.94VGPDD336 pKa = 2.69GRR338 pKa = 11.84LYY340 pKa = 10.73YY341 pKa = 9.88IYY343 pKa = 10.13EE344 pKa = 4.14EE345 pKa = 4.7TPGGPQLIGRR355 pKa = 11.84VNNPDD360 pKa = 3.6EE361 pKa = 4.59EE362 pKa = 4.42DD363 pKa = 3.93LEE365 pKa = 4.18EE366 pKa = 6.04LDD368 pKa = 4.98IEE370 pKa = 4.19EE371 pKa = 5.3DD372 pKa = 3.74PFNGTDD378 pKa = 3.4FCGRR382 pKa = 11.84VFPQFAPNQDD392 pKa = 3.1INPTVDD398 pKa = 5.18FDD400 pKa = 3.97WEE402 pKa = 4.27PDD404 pKa = 4.16FPCSNNPVQLTSQITPEE421 pKa = 3.8NYY423 pKa = 9.55RR424 pKa = 11.84PVSFEE429 pKa = 3.71WTFEE433 pKa = 4.05PPLTDD438 pKa = 3.35EE439 pKa = 5.27DD440 pKa = 4.69GEE442 pKa = 4.3PVDD445 pKa = 4.08IDD447 pKa = 3.81FNQEE451 pKa = 3.34HH452 pKa = 6.79LLIPEE457 pKa = 4.33EE458 pKa = 4.06ATANQSIQVTLTVTFADD475 pKa = 4.23GNVVTVPKK483 pKa = 9.91TIQLTEE489 pKa = 4.06NNLEE493 pKa = 4.42ANFSAQDD500 pKa = 3.42TTICEE505 pKa = 4.7GPCVDD510 pKa = 5.04IGSLLQVQEE519 pKa = 4.41GGGQGGEE526 pKa = 4.12NPNVGGPGQGGNYY539 pKa = 9.85EE540 pKa = 4.36YY541 pKa = 10.62FWSNRR546 pKa = 11.84RR547 pKa = 11.84DD548 pKa = 3.74EE549 pKa = 4.42GWVRR553 pKa = 11.84DD554 pKa = 3.56TDD556 pKa = 3.76NCVDD560 pKa = 3.66RR561 pKa = 11.84PGLYY565 pKa = 8.66WVLVRR570 pKa = 11.84EE571 pKa = 4.7PGSEE575 pKa = 3.97CYY577 pKa = 10.14AYY579 pKa = 10.63AEE581 pKa = 4.08VRR583 pKa = 11.84VRR585 pKa = 11.84IWDD588 pKa = 4.75LPDD591 pKa = 3.0QSNNVWLFGNGAGLDD606 pKa = 4.04FNPDD610 pKa = 3.58PDD612 pKa = 5.11DD613 pKa = 4.22EE614 pKa = 4.43EE615 pKa = 6.48APLPRR620 pKa = 11.84PVSHH624 pKa = 6.64NRR626 pKa = 11.84DD627 pKa = 3.09IPAGTTTISDD637 pKa = 3.26EE638 pKa = 4.24TGQVLFFTDD647 pKa = 5.18GEE649 pKa = 4.95SVWDD653 pKa = 4.04LNGDD657 pKa = 4.3LMANGDD663 pKa = 4.1NIGGNNQASEE673 pKa = 4.39GVIAVPVPQNQTIFYY688 pKa = 10.36VFTTQRR694 pKa = 11.84AADD697 pKa = 3.66GSNRR701 pKa = 11.84VSYY704 pKa = 10.91SVVDD708 pKa = 3.76IKK710 pKa = 10.38TDD712 pKa = 3.08NPTGVGSVVTKK723 pKa = 11.13NNFLFSPSTEE733 pKa = 3.72QTAALASGDD742 pKa = 3.95TTWVLFHH749 pKa = 7.06EE750 pKa = 5.11LGNNTFRR757 pKa = 11.84AYY759 pKa = 9.44PVSPFGIGSPVFSSVGSNHH778 pKa = 6.05NFTSGVGSMKK788 pKa = 10.45FSPDD792 pKa = 3.02GNQVAVTISDD802 pKa = 4.02GNCSRR807 pKa = 11.84LEE809 pKa = 3.56IFDD812 pKa = 4.87FNEE815 pKa = 3.57QTGRR819 pKa = 11.84LTEE822 pKa = 3.88YY823 pKa = 11.39ALIEE827 pKa = 4.34LGCGAEE833 pKa = 3.96EE834 pKa = 4.76VYY836 pKa = 10.94GLEE839 pKa = 4.09FSPDD843 pKa = 2.86GSRR846 pKa = 11.84IFVSYY851 pKa = 9.58TGSPGKK857 pKa = 9.98IEE859 pKa = 4.27EE860 pKa = 4.39YY861 pKa = 9.99LVKK864 pKa = 10.48RR865 pKa = 11.84PEE867 pKa = 4.1TAVEE871 pKa = 4.32GEE873 pKa = 5.19DD874 pKa = 3.28IDD876 pKa = 5.83SDD878 pKa = 4.48LSCFDD883 pKa = 3.85CFEE886 pKa = 4.34NASNRR891 pKa = 11.84SQRR894 pKa = 11.84EE895 pKa = 3.38NCIIDD900 pKa = 3.58SRR902 pKa = 11.84NVLSTAGPFGAIQIGPDD919 pKa = 3.28GQIYY923 pKa = 8.61VARR926 pKa = 11.84PDD928 pKa = 3.62RR929 pKa = 11.84NEE931 pKa = 3.36ITAILPGFNCLDD943 pKa = 3.25SFYY946 pKa = 10.96NEE948 pKa = 4.08QGVSLLPGTSMNLGLPAFVQQSGSSIPEE976 pKa = 3.67PALAGIEE983 pKa = 4.07RR984 pKa = 11.84LCFDD988 pKa = 4.43PDD990 pKa = 3.27GTIGLFEE997 pKa = 5.74AGGEE1001 pKa = 4.0PDD1003 pKa = 2.99IDD1005 pKa = 4.01SYY1007 pKa = 11.62FWTITHH1013 pKa = 6.97EE1014 pKa = 4.53DD1015 pKa = 3.91GEE1017 pKa = 5.16VVFEE1021 pKa = 4.24EE1022 pKa = 5.04GGPGEE1027 pKa = 4.34TFQEE1031 pKa = 4.24IEE1033 pKa = 4.02PLFPRR1038 pKa = 11.84VGIYY1042 pKa = 8.97TVALRR1047 pKa = 11.84VDD1049 pKa = 3.21RR1050 pKa = 11.84CGNPEE1055 pKa = 3.89YY1056 pKa = 10.37YY1057 pKa = 9.6RR1058 pKa = 11.84EE1059 pKa = 4.11SMEE1062 pKa = 4.19VEE1064 pKa = 4.64VIASPEE1070 pKa = 3.82LTLPEE1075 pKa = 4.35EE1076 pKa = 4.26ATLCVGTPVPLTAIDD1091 pKa = 4.93GYY1093 pKa = 11.26DD1094 pKa = 3.69PSEE1097 pKa = 3.87GLYY1100 pKa = 10.91DD1101 pKa = 3.92FEE1103 pKa = 4.48WRR1105 pKa = 11.84NAAGQLIGDD1114 pKa = 4.19EE1115 pKa = 4.15NSNTIEE1121 pKa = 4.03VVEE1124 pKa = 4.17EE1125 pKa = 3.83SVYY1128 pKa = 10.17TVTVRR1133 pKa = 11.84FRR1135 pKa = 11.84VPEE1138 pKa = 4.19GVDD1141 pKa = 3.2PEE1143 pKa = 4.78EE1144 pKa = 5.01FDD1146 pKa = 3.52ACPVSRR1152 pKa = 11.84SIFVGPAFDD1161 pKa = 4.66FDD1163 pKa = 5.56LIQSAEE1169 pKa = 4.3EE1170 pKa = 4.12VCLDD1174 pKa = 3.63EE1175 pKa = 4.6PVVIFSPNTPITGEE1189 pKa = 3.9WFFRR1193 pKa = 11.84PAGSPDD1199 pKa = 3.32RR1200 pKa = 11.84VSLGEE1205 pKa = 3.94FFEE1208 pKa = 5.64LEE1210 pKa = 4.9LIPASLPSPGDD1221 pKa = 3.3YY1222 pKa = 10.73EE1223 pKa = 4.79IIFFAEE1229 pKa = 4.27DD1230 pKa = 3.9PLVEE1234 pKa = 4.22GCFVEE1239 pKa = 5.65KK1240 pKa = 10.28IVPLTIFPLPAVDD1253 pKa = 5.73AVILSDD1259 pKa = 5.71AEE1261 pKa = 4.11DD1262 pKa = 4.29CEE1264 pKa = 4.73IPDD1267 pKa = 3.61GSFEE1271 pKa = 3.98ITALSDD1277 pKa = 3.45IEE1279 pKa = 4.19SLEE1282 pKa = 3.88VLEE1285 pKa = 5.39LGLLFGPIAAGEE1297 pKa = 4.07ALPPFTDD1304 pKa = 4.8LEE1306 pKa = 4.77PGLYY1310 pKa = 8.48TLQMLNGFGCEE1321 pKa = 4.77FIQSVTIRR1329 pKa = 11.84NLNPPRR1335 pKa = 11.84GIEE1338 pKa = 4.26GYY1340 pKa = 8.36EE1341 pKa = 3.87VEE1343 pKa = 4.92VSPEE1347 pKa = 3.63ICDD1350 pKa = 3.49VNGVLDD1356 pKa = 4.07GQLTIRR1362 pKa = 11.84FISGGASGTYY1372 pKa = 9.93QITRR1376 pKa = 11.84QGDD1379 pKa = 3.61GAFVSDD1385 pKa = 3.77NFNDD1389 pKa = 3.59VEE1391 pKa = 4.63EE1392 pKa = 4.34ITVPVPAGIYY1402 pKa = 9.39SVVVEE1407 pKa = 4.96DD1408 pKa = 4.16VDD1410 pKa = 4.07GCAVPFEE1417 pKa = 4.71DD1418 pKa = 3.78LVEE1421 pKa = 4.15VEE1423 pKa = 4.33EE1424 pKa = 4.35VEE1426 pKa = 4.37IVAFSAPGNVEE1437 pKa = 3.61ACEE1440 pKa = 4.29SFTFSPQSEE1449 pKa = 4.55DD1450 pKa = 3.25EE1451 pKa = 4.21LVYY1454 pKa = 9.84TLRR1457 pKa = 11.84DD1458 pKa = 3.42AQGNQIQATPEE1469 pKa = 3.89GSFTITEE1476 pKa = 4.04AGLYY1480 pKa = 9.91VLTGEE1485 pKa = 4.9DD1486 pKa = 3.8PDD1488 pKa = 4.8GIKK1491 pKa = 10.34CARR1494 pKa = 11.84EE1495 pKa = 3.54RR1496 pKa = 11.84EE1497 pKa = 4.35INVTLTTAPVFSLAGPRR1514 pKa = 11.84VDD1516 pKa = 4.57CDD1518 pKa = 3.3LGLFYY1523 pKa = 10.72EE1524 pKa = 4.97AVLGPNEE1531 pKa = 4.94DD1532 pKa = 3.92PNQVFIFWLDD1542 pKa = 3.45SNGEE1546 pKa = 4.33VVSRR1550 pKa = 11.84NPLFFPRR1557 pKa = 11.84VPGTYY1562 pKa = 9.67FLEE1565 pKa = 4.34VQPRR1569 pKa = 11.84TGSLCEE1575 pKa = 3.84IDD1577 pKa = 4.85PISFTVEE1584 pKa = 3.86DD1585 pKa = 4.62TDD1587 pKa = 3.8PQIDD1591 pKa = 3.97VEE1593 pKa = 4.88LEE1595 pKa = 3.81ALPFCAEE1602 pKa = 4.54DD1603 pKa = 3.94PFTLISVIADD1613 pKa = 3.84LSDD1616 pKa = 3.15VALIQWFEE1624 pKa = 4.17IINGEE1629 pKa = 4.29RR1630 pKa = 11.84EE1631 pKa = 4.64LIEE1634 pKa = 4.52DD1635 pKa = 4.85WIGLDD1640 pKa = 3.16QVVALFEE1647 pKa = 4.54GVFEE1651 pKa = 4.53IVLINNDD1658 pKa = 2.85GCEE1661 pKa = 3.76IGTAIIEE1668 pKa = 4.22IRR1670 pKa = 11.84QSTIGPPDD1678 pKa = 3.98LDD1680 pKa = 3.75EE1681 pKa = 6.07LYY1683 pKa = 10.79VICAPEE1689 pKa = 4.99GILVEE1694 pKa = 4.56IGPGEE1699 pKa = 3.95YY1700 pKa = 10.42DD1701 pKa = 3.11NYY1703 pKa = 10.86QWILDD1708 pKa = 4.02EE1709 pKa = 4.33EE1710 pKa = 4.9VVAVSPTFSPTIPGDD1725 pKa = 3.5YY1726 pKa = 10.48EE1727 pKa = 5.84LIVSDD1732 pKa = 5.62DD1733 pKa = 3.94LGCEE1737 pKa = 3.7FRR1739 pKa = 11.84ARR1741 pKa = 11.84FVVEE1745 pKa = 4.29EE1746 pKa = 4.09DD1747 pKa = 4.04CEE1749 pKa = 4.44LKK1751 pKa = 10.79INFPNAIIPNDD1762 pKa = 3.49SEE1764 pKa = 5.02RR1765 pKa = 11.84NFVVYY1770 pKa = 10.49TNDD1773 pKa = 3.31FVDD1776 pKa = 3.52EE1777 pKa = 4.74LEE1779 pKa = 4.17VFIYY1783 pKa = 10.45NRR1785 pKa = 11.84WGEE1788 pKa = 4.22LIFYY1792 pKa = 10.26CEE1794 pKa = 4.29SNGVNGEE1801 pKa = 4.05IPRR1804 pKa = 11.84CAWDD1808 pKa = 4.37GIVNGRR1814 pKa = 11.84TVPIGTYY1821 pKa = 9.46PVVVNYY1827 pKa = 10.51GSTSQNVRR1835 pKa = 11.84KK1836 pKa = 7.68TLKK1839 pKa = 9.52RR1840 pKa = 11.84TILVIEE1846 pKa = 4.2
Molecular weight: 201.82 kDa
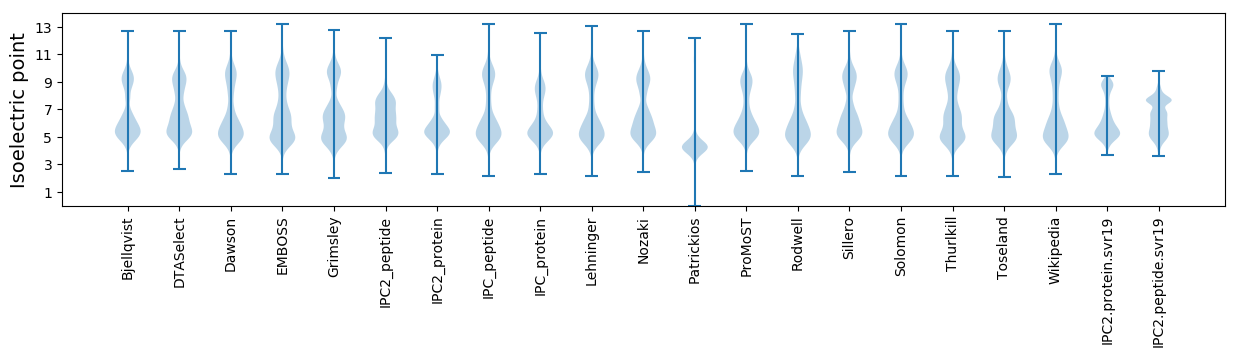
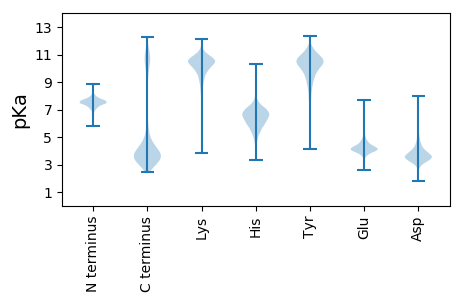
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K1LBU4|K1LBU4_9BACT Histidine protein kinase divJ OS=Cecembia lonarensis LW9 OX=1225176 GN=divJ_1 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.78HH16 pKa = 3.85GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VIKK32 pKa = 10.16SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84HH40 pKa = 5.41KK41 pKa = 10.43LTVSSEE47 pKa = 4.01KK48 pKa = 9.88TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.78HH16 pKa = 3.85GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VIKK32 pKa = 10.16SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84HH40 pKa = 5.41KK41 pKa = 10.43LTVSSEE47 pKa = 4.01KK48 pKa = 9.88TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1388209 |
29 |
2681 |
329.5 |
37.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.745 ± 0.035 | 0.688 ± 0.011 |
5.37 ± 0.029 | 7.001 ± 0.036 |
5.516 ± 0.034 | 7.07 ± 0.036 |
1.939 ± 0.018 | 7.283 ± 0.032 |
6.686 ± 0.043 | 9.903 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.53 ± 0.019 | 5.065 ± 0.033 |
3.99 ± 0.025 | 3.753 ± 0.021 |
4.293 ± 0.026 | 6.131 ± 0.025 |
4.798 ± 0.024 | 6.228 ± 0.03 |
1.289 ± 0.016 | 3.723 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
