
Yoonia tamlensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Yoonia
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
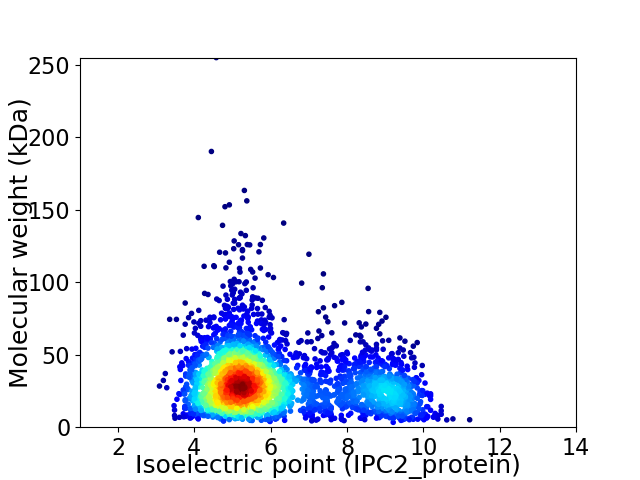
Virtual 2D-PAGE plot for 3187 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6HAJ4|A0A1I6HAJ4_9RHOB Putative peptidoglycan binding domain-containing protein OS=Yoonia tamlensis OX=390270 GN=SAMN04488005_2429 PE=4 SV=1
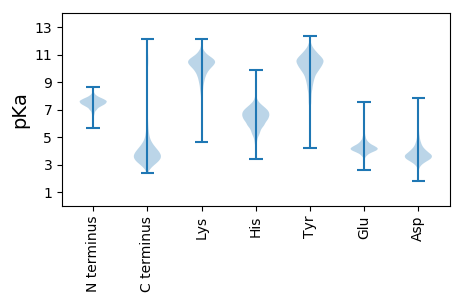
MM1 pKa = 7.2QFLNIRR7 pKa = 11.84SGALAVAMLGAAAPAVAQDD26 pKa = 4.48TYY28 pKa = 11.49CGGAGNGGQWIGGDD42 pKa = 3.62AQSSDD47 pKa = 2.79IATAGSYY54 pKa = 9.04QEE56 pKa = 4.45QMALVLGGNEE66 pKa = 4.21YY67 pKa = 10.4ISLFSLSQGTDD78 pKa = 3.04VRR80 pKa = 11.84IEE82 pKa = 3.79AAGRR86 pKa = 11.84GAGDD90 pKa = 3.85TIIDD94 pKa = 3.83VLDD97 pKa = 3.98SNGSIVASDD106 pKa = 4.27DD107 pKa = 3.88DD108 pKa = 4.38SGGNAASRR116 pKa = 11.84AEE118 pKa = 4.3TYY120 pKa = 10.87LEE122 pKa = 4.11AGTYY126 pKa = 10.45CMALRR131 pKa = 11.84SYY133 pKa = 11.27DD134 pKa = 4.11GGPMTAFVRR143 pKa = 11.84VGRR146 pKa = 11.84QEE148 pKa = 4.69HH149 pKa = 5.78EE150 pKa = 4.01ALTEE154 pKa = 4.02GVMDD158 pKa = 4.12SPVNDD163 pKa = 4.32GSAGGCATATPMGGLGSSVTGSVADD188 pKa = 3.66NPNWSFTLDD197 pKa = 3.25APAQISITAEE207 pKa = 3.89NEE209 pKa = 3.83SADD212 pKa = 3.85PVVTLYY218 pKa = 10.79GANDD222 pKa = 3.36MFIAEE227 pKa = 4.23NDD229 pKa = 4.02DD230 pKa = 3.72YY231 pKa = 11.92DD232 pKa = 4.15GLNSRR237 pKa = 11.84IDD239 pKa = 3.48VTDD242 pKa = 3.27TLDD245 pKa = 3.72PGTYY249 pKa = 9.25CIAMAALSDD258 pKa = 4.07EE259 pKa = 4.45YY260 pKa = 11.76APITVSLTEE269 pKa = 4.28YY270 pKa = 10.69DD271 pKa = 3.81PEE273 pKa = 5.26AATLSLYY280 pKa = 10.76DD281 pKa = 4.05RR282 pKa = 11.84GEE284 pKa = 4.05AAPPLDD290 pKa = 3.38GSIPVTDD297 pKa = 4.46LGVLEE302 pKa = 4.28SRR304 pKa = 11.84VRR306 pKa = 11.84QDD308 pKa = 2.85VQATDD313 pKa = 3.34DD314 pKa = 4.05VTWFTLEE321 pKa = 3.99VLDD324 pKa = 5.02NSLLVIEE331 pKa = 5.58AIGGGNSDD339 pKa = 3.6PWLVVYY345 pKa = 10.62DD346 pKa = 3.91DD347 pKa = 4.61LGRR350 pKa = 11.84EE351 pKa = 3.83VGMNDD356 pKa = 4.42DD357 pKa = 4.47YY358 pKa = 11.99GDD360 pKa = 4.2GLDD363 pKa = 3.81SLVMARR369 pKa = 11.84VQSGTYY375 pKa = 9.44IVGVRR380 pKa = 11.84QFDD383 pKa = 3.6NTAGLIRR390 pKa = 11.84LLAEE394 pKa = 4.12RR395 pKa = 11.84YY396 pKa = 9.25VRR398 pKa = 11.84AQQ400 pKa = 2.93
MM1 pKa = 7.2QFLNIRR7 pKa = 11.84SGALAVAMLGAAAPAVAQDD26 pKa = 4.48TYY28 pKa = 11.49CGGAGNGGQWIGGDD42 pKa = 3.62AQSSDD47 pKa = 2.79IATAGSYY54 pKa = 9.04QEE56 pKa = 4.45QMALVLGGNEE66 pKa = 4.21YY67 pKa = 10.4ISLFSLSQGTDD78 pKa = 3.04VRR80 pKa = 11.84IEE82 pKa = 3.79AAGRR86 pKa = 11.84GAGDD90 pKa = 3.85TIIDD94 pKa = 3.83VLDD97 pKa = 3.98SNGSIVASDD106 pKa = 4.27DD107 pKa = 3.88DD108 pKa = 4.38SGGNAASRR116 pKa = 11.84AEE118 pKa = 4.3TYY120 pKa = 10.87LEE122 pKa = 4.11AGTYY126 pKa = 10.45CMALRR131 pKa = 11.84SYY133 pKa = 11.27DD134 pKa = 4.11GGPMTAFVRR143 pKa = 11.84VGRR146 pKa = 11.84QEE148 pKa = 4.69HH149 pKa = 5.78EE150 pKa = 4.01ALTEE154 pKa = 4.02GVMDD158 pKa = 4.12SPVNDD163 pKa = 4.32GSAGGCATATPMGGLGSSVTGSVADD188 pKa = 3.66NPNWSFTLDD197 pKa = 3.25APAQISITAEE207 pKa = 3.89NEE209 pKa = 3.83SADD212 pKa = 3.85PVVTLYY218 pKa = 10.79GANDD222 pKa = 3.36MFIAEE227 pKa = 4.23NDD229 pKa = 4.02DD230 pKa = 3.72YY231 pKa = 11.92DD232 pKa = 4.15GLNSRR237 pKa = 11.84IDD239 pKa = 3.48VTDD242 pKa = 3.27TLDD245 pKa = 3.72PGTYY249 pKa = 9.25CIAMAALSDD258 pKa = 4.07EE259 pKa = 4.45YY260 pKa = 11.76APITVSLTEE269 pKa = 4.28YY270 pKa = 10.69DD271 pKa = 3.81PEE273 pKa = 5.26AATLSLYY280 pKa = 10.76DD281 pKa = 4.05RR282 pKa = 11.84GEE284 pKa = 4.05AAPPLDD290 pKa = 3.38GSIPVTDD297 pKa = 4.46LGVLEE302 pKa = 4.28SRR304 pKa = 11.84VRR306 pKa = 11.84QDD308 pKa = 2.85VQATDD313 pKa = 3.34DD314 pKa = 4.05VTWFTLEE321 pKa = 3.99VLDD324 pKa = 5.02NSLLVIEE331 pKa = 5.58AIGGGNSDD339 pKa = 3.6PWLVVYY345 pKa = 10.62DD346 pKa = 3.91DD347 pKa = 4.61LGRR350 pKa = 11.84EE351 pKa = 3.83VGMNDD356 pKa = 4.42DD357 pKa = 4.47YY358 pKa = 11.99GDD360 pKa = 4.2GLDD363 pKa = 3.81SLVMARR369 pKa = 11.84VQSGTYY375 pKa = 9.44IVGVRR380 pKa = 11.84QFDD383 pKa = 3.6NTAGLIRR390 pKa = 11.84LLAEE394 pKa = 4.12RR395 pKa = 11.84YY396 pKa = 9.25VRR398 pKa = 11.84AQQ400 pKa = 2.93
Molecular weight: 41.71 kDa
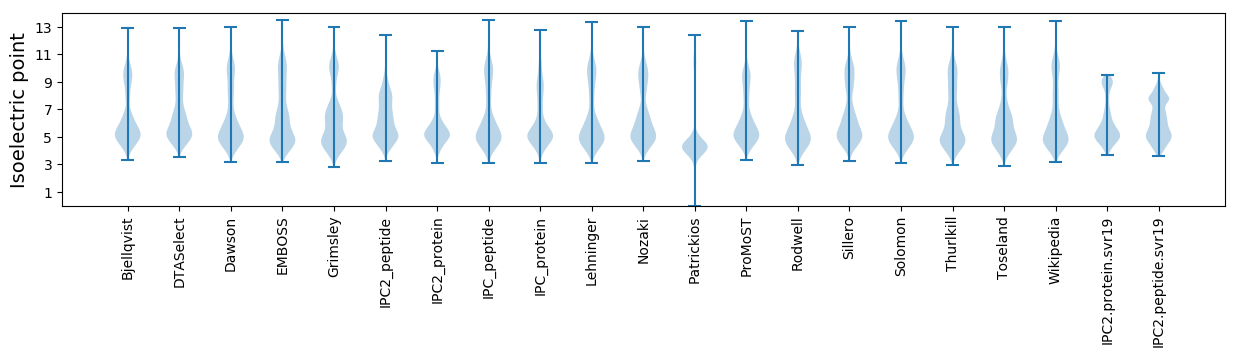
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6FQ09|A0A1I6FQ09_9RHOB Nucleobase:cation symporter-2 NCS2 family OS=Yoonia tamlensis OX=390270 GN=SAMN04488005_0233 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.53ILNNRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.47GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.53ILNNRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.47GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
974354 |
31 |
2420 |
305.7 |
33.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.616 ± 0.066 | 0.933 ± 0.012 |
6.339 ± 0.03 | 5.022 ± 0.042 |
3.856 ± 0.031 | 8.434 ± 0.04 |
2.074 ± 0.025 | 5.76 ± 0.033 |
3.32 ± 0.039 | 9.665 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.868 ± 0.023 | 2.946 ± 0.023 |
4.86 ± 0.03 | 3.554 ± 0.024 |
6.003 ± 0.041 | 5.001 ± 0.031 |
5.813 ± 0.036 | 7.257 ± 0.035 |
1.344 ± 0.016 | 2.336 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
