
Roseiconus nitratireducens
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Roseiconus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
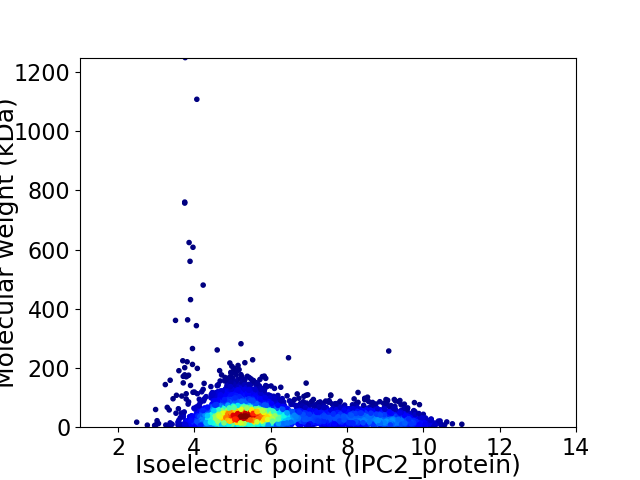
Virtual 2D-PAGE plot for 5599 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
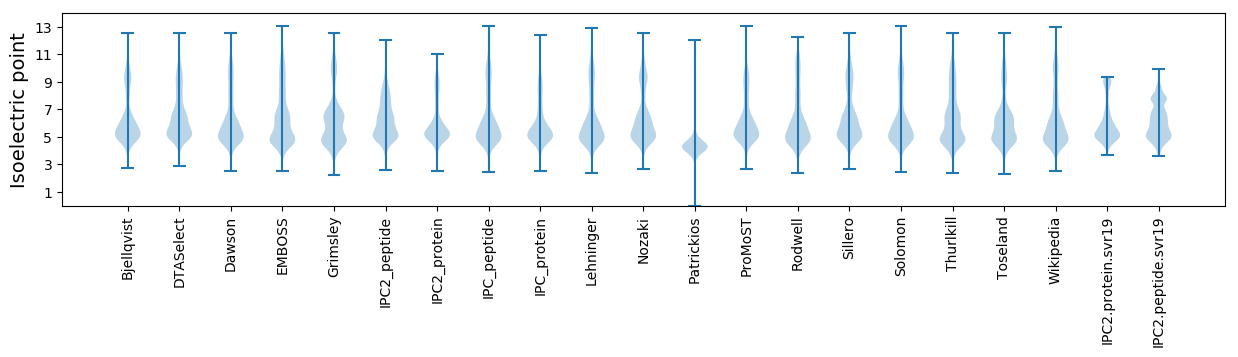
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5M6D3T7|A0A5M6D3T7_9BACT Glycosyl transferase family 25 OS=Roseiconus nitratireducens OX=2605748 GN=FYK55_18440 PE=4 SV=1
MM1 pKa = 7.37FPNPPGRR8 pKa = 11.84FAQSRR13 pKa = 11.84YY14 pKa = 9.03RR15 pKa = 11.84CLTRR19 pKa = 11.84GLEE22 pKa = 4.02NLEE25 pKa = 4.01RR26 pKa = 11.84RR27 pKa = 11.84CLLSFDD33 pKa = 4.2YY34 pKa = 11.42ASMVQVAPGLAVDD47 pKa = 4.1PGDD50 pKa = 3.86YY51 pKa = 11.03EE52 pKa = 4.22EE53 pKa = 5.83GSLLVKK59 pKa = 10.36YY60 pKa = 9.82KK61 pKa = 8.92QTGIGAVRR69 pKa = 11.84VTEE72 pKa = 4.17SLGATVGRR80 pKa = 11.84AYY82 pKa = 9.5TQIPGLHH89 pKa = 4.82QVKK92 pKa = 10.08LPSGVKK98 pKa = 10.0VADD101 pKa = 3.28VLPRR105 pKa = 11.84FRR107 pKa = 11.84NNPNVLYY114 pKa = 10.67AEE116 pKa = 4.5PDD118 pKa = 3.54FRR120 pKa = 11.84LHH122 pKa = 5.27TTALPNDD129 pKa = 3.59PRR131 pKa = 11.84YY132 pKa = 10.83DD133 pKa = 3.96EE134 pKa = 5.42LWGLNNTGQTGGTVDD149 pKa = 4.04ADD151 pKa = 3.66SDD153 pKa = 3.66IAEE156 pKa = 4.44AWDD159 pKa = 3.23IATGSNLVVAVIDD172 pKa = 3.55TGVDD176 pKa = 3.51YY177 pKa = 11.29LHH179 pKa = 7.45EE180 pKa = 4.87DD181 pKa = 3.38LAANMWVNAGEE192 pKa = 4.17IPGDD196 pKa = 4.39GIDD199 pKa = 3.47NDD201 pKa = 4.24GNGFVDD207 pKa = 6.01DD208 pKa = 3.28IHH210 pKa = 8.6GYY212 pKa = 10.82DD213 pKa = 4.63FANDD217 pKa = 4.4DD218 pKa = 4.89PDD220 pKa = 5.01PMDD223 pKa = 4.27DD224 pKa = 3.65QGHH227 pKa = 5.39GTHH230 pKa = 6.09VAGTIGAVGDD240 pKa = 3.65NGIGVVGVNWDD251 pKa = 3.63VQLMAVKK258 pKa = 10.32FLDD261 pKa = 3.39ATGNGTTSDD270 pKa = 3.45AVDD273 pKa = 4.83AINYY277 pKa = 8.56AVANGAQIANASWGGNEE294 pKa = 4.44PFSQALYY301 pKa = 10.63DD302 pKa = 4.79AIAGGRR308 pKa = 11.84DD309 pKa = 2.87AGQIFIAGAGNGNAFGFGQDD329 pKa = 3.09NDD331 pKa = 4.31SNPFYY336 pKa = 10.12PASYY340 pKa = 10.72DD341 pKa = 3.34LDD343 pKa = 4.46NIISVAATDD352 pKa = 4.25HH353 pKa = 6.16NDD355 pKa = 2.87ARR357 pKa = 11.84ATFSNYY363 pKa = 10.51GITSVDD369 pKa = 3.3LAGPGVSILSTRR381 pKa = 11.84PGNSYY386 pKa = 10.5EE387 pKa = 4.16LLSGTSMATPHH398 pKa = 5.53VAGVASLVWQHH409 pKa = 5.67NPTWTADD416 pKa = 3.51QVIEE420 pKa = 4.28QILNSVDD427 pKa = 3.81PVSSMQGITTTGGRR441 pKa = 11.84LNAAAALGEE450 pKa = 4.5PVPPPPPPPPATLPVSEE467 pKa = 5.15DD468 pKa = 3.57FEE470 pKa = 5.11DD471 pKa = 5.33ALAQDD476 pKa = 3.78FQVRR480 pKa = 11.84VGTWGVTSGRR490 pKa = 11.84YY491 pKa = 9.64AITPVMGDD499 pKa = 3.62PSASAVSTVRR509 pKa = 11.84LEE511 pKa = 4.15TPLADD516 pKa = 3.99DD517 pKa = 4.1MEE519 pKa = 4.76VFATANAAPEE529 pKa = 3.66IGFFFNYY536 pKa = 10.01LSNGYY541 pKa = 9.45IVFDD545 pKa = 3.76YY546 pKa = 11.07QDD548 pKa = 3.56NFDD551 pKa = 3.77FKK553 pKa = 10.7YY554 pKa = 10.72AGFNVVSGQWVIGQRR569 pKa = 11.84SGSGWQNLATLSDD582 pKa = 3.54AGIAASTDD590 pKa = 2.59VGMRR594 pKa = 11.84LTIEE598 pKa = 4.92DD599 pKa = 4.09GQSVTLYY606 pKa = 11.01GKK608 pKa = 9.3GAPVLSHH615 pKa = 6.59SFAEE619 pKa = 4.45SVVDD623 pKa = 4.02GQVGVGAQNSQTAFDD638 pKa = 3.68NVYY641 pKa = 10.75VNTFVSATPAALPIQEE657 pKa = 4.96DD658 pKa = 4.29FSDD661 pKa = 3.84GTADD665 pKa = 3.33NFQPQTPGWSVSGGAYY681 pKa = 9.61QVDD684 pKa = 4.24AFTGANAFSTLQVDD698 pKa = 4.71GALPADD704 pKa = 3.76LEE706 pKa = 4.34LRR708 pKa = 11.84ATANVHH714 pKa = 6.1APSGSQSSNALLVFDD729 pKa = 4.61YY730 pKa = 11.21HH731 pKa = 7.85NEE733 pKa = 3.79NDD735 pKa = 3.86FKK737 pKa = 11.17FVAAYY742 pKa = 10.17AGTDD746 pKa = 2.98QWVIGVRR753 pKa = 11.84GAGWTTVAEE762 pKa = 4.05TSEE765 pKa = 4.17AVEE768 pKa = 4.34TNVDD772 pKa = 2.66YY773 pKa = 11.6DD774 pKa = 3.76MQVVIEE780 pKa = 5.08DD781 pKa = 4.09GSSVTLLVDD790 pKa = 3.99GVVKK794 pKa = 9.84ATHH797 pKa = 6.52TFADD801 pKa = 3.9SVTDD805 pKa = 4.02GVLGLGTLNASVHH818 pKa = 6.12FDD820 pKa = 3.38DD821 pKa = 6.62LVVQEE826 pKa = 5.08YY827 pKa = 10.37LPPPPPPPGSLPIVEE842 pKa = 5.0DD843 pKa = 4.41FSDD846 pKa = 3.72GLADD850 pKa = 3.65YY851 pKa = 7.53FQPQLGNWSVNNGQYY866 pKa = 10.33FGSPSGDD873 pKa = 3.09ALSTLNLAEE882 pKa = 4.96ALPSDD887 pKa = 4.06LDD889 pKa = 3.59FRR891 pKa = 11.84ATVAVQQPTSGLYY904 pKa = 9.89QNAVMIFDD912 pKa = 4.0YY913 pKa = 10.94QSPTNFKK920 pKa = 10.41FAGAYY925 pKa = 10.1AGTDD929 pKa = 2.91QWVIGQRR936 pKa = 11.84TSQWVTLASAGAAVDD951 pKa = 3.51VGVDD955 pKa = 3.2YY956 pKa = 11.1DD957 pKa = 3.88LRR959 pKa = 11.84LVIQAGNQVALYY971 pKa = 10.91ANGALQTTYY980 pKa = 11.06AFSGSVTDD988 pKa = 4.04GAIGLGTWNALARR1001 pKa = 11.84FDD1003 pKa = 5.14DD1004 pKa = 4.25VSLQTYY1010 pKa = 7.31VAPPPPPAASLPLSEE1025 pKa = 5.39DD1026 pKa = 3.47FSDD1029 pKa = 4.22GVADD1033 pKa = 4.83FFQPQVGDD1041 pKa = 3.43WLVNAGRR1048 pKa = 11.84YY1049 pKa = 8.5QGSPSGDD1056 pKa = 2.91AVTTLNVAEE1065 pKa = 4.52PLPTEE1070 pKa = 4.15LEE1072 pKa = 4.07FRR1074 pKa = 11.84ASVTVHH1080 pKa = 5.94PPAYY1084 pKa = 10.08GFYY1087 pKa = 10.62QNGLLIFDD1095 pKa = 4.25YY1096 pKa = 10.92QSPTNFKK1103 pKa = 10.41FAGAYY1108 pKa = 10.1AGTDD1112 pKa = 2.94QWVIGHH1118 pKa = 6.3RR1119 pKa = 11.84TSQWITDD1126 pKa = 3.63ASAGASIDD1134 pKa = 3.36VGTDD1138 pKa = 3.16YY1139 pKa = 11.05EE1140 pKa = 4.43LSVVIEE1146 pKa = 4.48GDD1148 pKa = 3.56HH1149 pKa = 6.08QVTLLVDD1156 pKa = 4.07GVPQTSHH1163 pKa = 6.16TFSDD1167 pKa = 3.81SLTDD1171 pKa = 3.44GDD1173 pKa = 5.28LGFGTWNAVASYY1185 pKa = 11.09DD1186 pKa = 3.86DD1187 pKa = 4.49VSVQSYY1193 pKa = 7.67VAPPPPPPPPAGTLPVAEE1211 pKa = 5.05DD1212 pKa = 4.1FSDD1215 pKa = 4.19GVADD1219 pKa = 3.65YY1220 pKa = 7.93FQPQVGDD1227 pKa = 3.29WAINAGRR1234 pKa = 11.84YY1235 pKa = 9.14VGTPAGDD1242 pKa = 3.56AVSTLALDD1250 pKa = 4.08APLPSDD1256 pKa = 4.94LEE1258 pKa = 4.16FQATVNIQPPSGGLYY1273 pKa = 10.35QNGLLVFDD1281 pKa = 4.51YY1282 pKa = 10.88QSPTDD1287 pKa = 3.85FKK1289 pKa = 11.29FAGAYY1294 pKa = 10.12AGTNQWVIGHH1304 pKa = 6.21RR1305 pKa = 11.84TSQWVTDD1312 pKa = 4.03AFAGSTVNVGVDD1324 pKa = 3.15YY1325 pKa = 11.0DD1326 pKa = 3.81LRR1328 pKa = 11.84LVIEE1332 pKa = 5.02GGNQVTLWVGEE1343 pKa = 4.53TPQVTYY1349 pKa = 11.15SFAEE1353 pKa = 4.18SVTDD1357 pKa = 3.82GDD1359 pKa = 4.3IGLGTWNAVAKK1370 pKa = 10.01YY1371 pKa = 10.08DD1372 pKa = 3.9QIVVQAYY1379 pKa = 6.74TPPPPPPAAEE1389 pKa = 4.35LPVQEE1394 pKa = 5.45DD1395 pKa = 4.25YY1396 pKa = 11.82SDD1398 pKa = 3.73GLADD1402 pKa = 3.95LFVPRR1407 pKa = 11.84RR1408 pKa = 11.84GTWTVSGGEE1417 pKa = 4.11YY1418 pKa = 9.79HH1419 pKa = 7.53ASPSGDD1425 pKa = 3.54AISTLEE1431 pKa = 4.34PGGALPTDD1439 pKa = 4.31LEE1441 pKa = 4.47FQATARR1447 pKa = 11.84IQPPTYY1453 pKa = 10.22GVYY1456 pKa = 10.44QNAVVIFDD1464 pKa = 3.92YY1465 pKa = 11.01QSPTNFKK1472 pKa = 10.41FAGAYY1477 pKa = 10.1AGTNEE1482 pKa = 4.07WVIGTRR1488 pKa = 11.84TSSWTVLASYY1498 pKa = 11.3GEE1500 pKa = 4.46TVNVNVDD1507 pKa = 3.14YY1508 pKa = 10.39TLRR1511 pKa = 11.84LVIDD1515 pKa = 4.53GDD1517 pKa = 3.87HH1518 pKa = 5.99QATLFVDD1525 pKa = 4.54GLPKK1529 pKa = 8.5TTHH1532 pKa = 5.91TFNGSLTDD1540 pKa = 3.87GSVGLATWNAIAHH1553 pKa = 6.37FDD1555 pKa = 4.23DD1556 pKa = 4.91VSLQATGTSVAALTVGTSVLPEE1578 pKa = 4.06TNADD1582 pKa = 3.55LSDD1585 pKa = 4.43PLLSLPAASSRR1596 pKa = 11.84RR1597 pKa = 11.84SSAGIGASPSDD1608 pKa = 3.75TSAGAASHH1616 pKa = 6.2GRR1618 pKa = 11.84GNGRR1622 pKa = 11.84PGSPQSISSEE1632 pKa = 4.25SPLDD1636 pKa = 3.47QVFAEE1641 pKa = 4.79LEE1643 pKa = 3.89FDD1645 pKa = 5.01LMGDD1649 pKa = 4.12LSS1651 pKa = 3.72
MM1 pKa = 7.37FPNPPGRR8 pKa = 11.84FAQSRR13 pKa = 11.84YY14 pKa = 9.03RR15 pKa = 11.84CLTRR19 pKa = 11.84GLEE22 pKa = 4.02NLEE25 pKa = 4.01RR26 pKa = 11.84RR27 pKa = 11.84CLLSFDD33 pKa = 4.2YY34 pKa = 11.42ASMVQVAPGLAVDD47 pKa = 4.1PGDD50 pKa = 3.86YY51 pKa = 11.03EE52 pKa = 4.22EE53 pKa = 5.83GSLLVKK59 pKa = 10.36YY60 pKa = 9.82KK61 pKa = 8.92QTGIGAVRR69 pKa = 11.84VTEE72 pKa = 4.17SLGATVGRR80 pKa = 11.84AYY82 pKa = 9.5TQIPGLHH89 pKa = 4.82QVKK92 pKa = 10.08LPSGVKK98 pKa = 10.0VADD101 pKa = 3.28VLPRR105 pKa = 11.84FRR107 pKa = 11.84NNPNVLYY114 pKa = 10.67AEE116 pKa = 4.5PDD118 pKa = 3.54FRR120 pKa = 11.84LHH122 pKa = 5.27TTALPNDD129 pKa = 3.59PRR131 pKa = 11.84YY132 pKa = 10.83DD133 pKa = 3.96EE134 pKa = 5.42LWGLNNTGQTGGTVDD149 pKa = 4.04ADD151 pKa = 3.66SDD153 pKa = 3.66IAEE156 pKa = 4.44AWDD159 pKa = 3.23IATGSNLVVAVIDD172 pKa = 3.55TGVDD176 pKa = 3.51YY177 pKa = 11.29LHH179 pKa = 7.45EE180 pKa = 4.87DD181 pKa = 3.38LAANMWVNAGEE192 pKa = 4.17IPGDD196 pKa = 4.39GIDD199 pKa = 3.47NDD201 pKa = 4.24GNGFVDD207 pKa = 6.01DD208 pKa = 3.28IHH210 pKa = 8.6GYY212 pKa = 10.82DD213 pKa = 4.63FANDD217 pKa = 4.4DD218 pKa = 4.89PDD220 pKa = 5.01PMDD223 pKa = 4.27DD224 pKa = 3.65QGHH227 pKa = 5.39GTHH230 pKa = 6.09VAGTIGAVGDD240 pKa = 3.65NGIGVVGVNWDD251 pKa = 3.63VQLMAVKK258 pKa = 10.32FLDD261 pKa = 3.39ATGNGTTSDD270 pKa = 3.45AVDD273 pKa = 4.83AINYY277 pKa = 8.56AVANGAQIANASWGGNEE294 pKa = 4.44PFSQALYY301 pKa = 10.63DD302 pKa = 4.79AIAGGRR308 pKa = 11.84DD309 pKa = 2.87AGQIFIAGAGNGNAFGFGQDD329 pKa = 3.09NDD331 pKa = 4.31SNPFYY336 pKa = 10.12PASYY340 pKa = 10.72DD341 pKa = 3.34LDD343 pKa = 4.46NIISVAATDD352 pKa = 4.25HH353 pKa = 6.16NDD355 pKa = 2.87ARR357 pKa = 11.84ATFSNYY363 pKa = 10.51GITSVDD369 pKa = 3.3LAGPGVSILSTRR381 pKa = 11.84PGNSYY386 pKa = 10.5EE387 pKa = 4.16LLSGTSMATPHH398 pKa = 5.53VAGVASLVWQHH409 pKa = 5.67NPTWTADD416 pKa = 3.51QVIEE420 pKa = 4.28QILNSVDD427 pKa = 3.81PVSSMQGITTTGGRR441 pKa = 11.84LNAAAALGEE450 pKa = 4.5PVPPPPPPPPATLPVSEE467 pKa = 5.15DD468 pKa = 3.57FEE470 pKa = 5.11DD471 pKa = 5.33ALAQDD476 pKa = 3.78FQVRR480 pKa = 11.84VGTWGVTSGRR490 pKa = 11.84YY491 pKa = 9.64AITPVMGDD499 pKa = 3.62PSASAVSTVRR509 pKa = 11.84LEE511 pKa = 4.15TPLADD516 pKa = 3.99DD517 pKa = 4.1MEE519 pKa = 4.76VFATANAAPEE529 pKa = 3.66IGFFFNYY536 pKa = 10.01LSNGYY541 pKa = 9.45IVFDD545 pKa = 3.76YY546 pKa = 11.07QDD548 pKa = 3.56NFDD551 pKa = 3.77FKK553 pKa = 10.7YY554 pKa = 10.72AGFNVVSGQWVIGQRR569 pKa = 11.84SGSGWQNLATLSDD582 pKa = 3.54AGIAASTDD590 pKa = 2.59VGMRR594 pKa = 11.84LTIEE598 pKa = 4.92DD599 pKa = 4.09GQSVTLYY606 pKa = 11.01GKK608 pKa = 9.3GAPVLSHH615 pKa = 6.59SFAEE619 pKa = 4.45SVVDD623 pKa = 4.02GQVGVGAQNSQTAFDD638 pKa = 3.68NVYY641 pKa = 10.75VNTFVSATPAALPIQEE657 pKa = 4.96DD658 pKa = 4.29FSDD661 pKa = 3.84GTADD665 pKa = 3.33NFQPQTPGWSVSGGAYY681 pKa = 9.61QVDD684 pKa = 4.24AFTGANAFSTLQVDD698 pKa = 4.71GALPADD704 pKa = 3.76LEE706 pKa = 4.34LRR708 pKa = 11.84ATANVHH714 pKa = 6.1APSGSQSSNALLVFDD729 pKa = 4.61YY730 pKa = 11.21HH731 pKa = 7.85NEE733 pKa = 3.79NDD735 pKa = 3.86FKK737 pKa = 11.17FVAAYY742 pKa = 10.17AGTDD746 pKa = 2.98QWVIGVRR753 pKa = 11.84GAGWTTVAEE762 pKa = 4.05TSEE765 pKa = 4.17AVEE768 pKa = 4.34TNVDD772 pKa = 2.66YY773 pKa = 11.6DD774 pKa = 3.76MQVVIEE780 pKa = 5.08DD781 pKa = 4.09GSSVTLLVDD790 pKa = 3.99GVVKK794 pKa = 9.84ATHH797 pKa = 6.52TFADD801 pKa = 3.9SVTDD805 pKa = 4.02GVLGLGTLNASVHH818 pKa = 6.12FDD820 pKa = 3.38DD821 pKa = 6.62LVVQEE826 pKa = 5.08YY827 pKa = 10.37LPPPPPPPGSLPIVEE842 pKa = 5.0DD843 pKa = 4.41FSDD846 pKa = 3.72GLADD850 pKa = 3.65YY851 pKa = 7.53FQPQLGNWSVNNGQYY866 pKa = 10.33FGSPSGDD873 pKa = 3.09ALSTLNLAEE882 pKa = 4.96ALPSDD887 pKa = 4.06LDD889 pKa = 3.59FRR891 pKa = 11.84ATVAVQQPTSGLYY904 pKa = 9.89QNAVMIFDD912 pKa = 4.0YY913 pKa = 10.94QSPTNFKK920 pKa = 10.41FAGAYY925 pKa = 10.1AGTDD929 pKa = 2.91QWVIGQRR936 pKa = 11.84TSQWVTLASAGAAVDD951 pKa = 3.51VGVDD955 pKa = 3.2YY956 pKa = 11.1DD957 pKa = 3.88LRR959 pKa = 11.84LVIQAGNQVALYY971 pKa = 10.91ANGALQTTYY980 pKa = 11.06AFSGSVTDD988 pKa = 4.04GAIGLGTWNALARR1001 pKa = 11.84FDD1003 pKa = 5.14DD1004 pKa = 4.25VSLQTYY1010 pKa = 7.31VAPPPPPAASLPLSEE1025 pKa = 5.39DD1026 pKa = 3.47FSDD1029 pKa = 4.22GVADD1033 pKa = 4.83FFQPQVGDD1041 pKa = 3.43WLVNAGRR1048 pKa = 11.84YY1049 pKa = 8.5QGSPSGDD1056 pKa = 2.91AVTTLNVAEE1065 pKa = 4.52PLPTEE1070 pKa = 4.15LEE1072 pKa = 4.07FRR1074 pKa = 11.84ASVTVHH1080 pKa = 5.94PPAYY1084 pKa = 10.08GFYY1087 pKa = 10.62QNGLLIFDD1095 pKa = 4.25YY1096 pKa = 10.92QSPTNFKK1103 pKa = 10.41FAGAYY1108 pKa = 10.1AGTDD1112 pKa = 2.94QWVIGHH1118 pKa = 6.3RR1119 pKa = 11.84TSQWITDD1126 pKa = 3.63ASAGASIDD1134 pKa = 3.36VGTDD1138 pKa = 3.16YY1139 pKa = 11.05EE1140 pKa = 4.43LSVVIEE1146 pKa = 4.48GDD1148 pKa = 3.56HH1149 pKa = 6.08QVTLLVDD1156 pKa = 4.07GVPQTSHH1163 pKa = 6.16TFSDD1167 pKa = 3.81SLTDD1171 pKa = 3.44GDD1173 pKa = 5.28LGFGTWNAVASYY1185 pKa = 11.09DD1186 pKa = 3.86DD1187 pKa = 4.49VSVQSYY1193 pKa = 7.67VAPPPPPPPPAGTLPVAEE1211 pKa = 5.05DD1212 pKa = 4.1FSDD1215 pKa = 4.19GVADD1219 pKa = 3.65YY1220 pKa = 7.93FQPQVGDD1227 pKa = 3.29WAINAGRR1234 pKa = 11.84YY1235 pKa = 9.14VGTPAGDD1242 pKa = 3.56AVSTLALDD1250 pKa = 4.08APLPSDD1256 pKa = 4.94LEE1258 pKa = 4.16FQATVNIQPPSGGLYY1273 pKa = 10.35QNGLLVFDD1281 pKa = 4.51YY1282 pKa = 10.88QSPTDD1287 pKa = 3.85FKK1289 pKa = 11.29FAGAYY1294 pKa = 10.12AGTNQWVIGHH1304 pKa = 6.21RR1305 pKa = 11.84TSQWVTDD1312 pKa = 4.03AFAGSTVNVGVDD1324 pKa = 3.15YY1325 pKa = 11.0DD1326 pKa = 3.81LRR1328 pKa = 11.84LVIEE1332 pKa = 5.02GGNQVTLWVGEE1343 pKa = 4.53TPQVTYY1349 pKa = 11.15SFAEE1353 pKa = 4.18SVTDD1357 pKa = 3.82GDD1359 pKa = 4.3IGLGTWNAVAKK1370 pKa = 10.01YY1371 pKa = 10.08DD1372 pKa = 3.9QIVVQAYY1379 pKa = 6.74TPPPPPPAAEE1389 pKa = 4.35LPVQEE1394 pKa = 5.45DD1395 pKa = 4.25YY1396 pKa = 11.82SDD1398 pKa = 3.73GLADD1402 pKa = 3.95LFVPRR1407 pKa = 11.84RR1408 pKa = 11.84GTWTVSGGEE1417 pKa = 4.11YY1418 pKa = 9.79HH1419 pKa = 7.53ASPSGDD1425 pKa = 3.54AISTLEE1431 pKa = 4.34PGGALPTDD1439 pKa = 4.31LEE1441 pKa = 4.47FQATARR1447 pKa = 11.84IQPPTYY1453 pKa = 10.22GVYY1456 pKa = 10.44QNAVVIFDD1464 pKa = 3.92YY1465 pKa = 11.01QSPTNFKK1472 pKa = 10.41FAGAYY1477 pKa = 10.1AGTNEE1482 pKa = 4.07WVIGTRR1488 pKa = 11.84TSSWTVLASYY1498 pKa = 11.3GEE1500 pKa = 4.46TVNVNVDD1507 pKa = 3.14YY1508 pKa = 10.39TLRR1511 pKa = 11.84LVIDD1515 pKa = 4.53GDD1517 pKa = 3.87HH1518 pKa = 5.99QATLFVDD1525 pKa = 4.54GLPKK1529 pKa = 8.5TTHH1532 pKa = 5.91TFNGSLTDD1540 pKa = 3.87GSVGLATWNAIAHH1553 pKa = 6.37FDD1555 pKa = 4.23DD1556 pKa = 4.91VSLQATGTSVAALTVGTSVLPEE1578 pKa = 4.06TNADD1582 pKa = 3.55LSDD1585 pKa = 4.43PLLSLPAASSRR1596 pKa = 11.84RR1597 pKa = 11.84SSAGIGASPSDD1608 pKa = 3.75TSAGAASHH1616 pKa = 6.2GRR1618 pKa = 11.84GNGRR1622 pKa = 11.84PGSPQSISSEE1632 pKa = 4.25SPLDD1636 pKa = 3.47QVFAEE1641 pKa = 4.79LEE1643 pKa = 3.89FDD1645 pKa = 5.01LMGDD1649 pKa = 4.12LSS1651 pKa = 3.72
Molecular weight: 173.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5M6DFK1|A0A5M6DFK1_9BACT Uncharacterized protein OS=Roseiconus nitratireducens OX=2605748 GN=FYK55_04130 PE=4 SV=1
MM1 pKa = 7.23SRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84SPLAAHH11 pKa = 6.89HH12 pKa = 6.6SPLTTRR18 pKa = 11.84HH19 pKa = 5.83SPLTTHH25 pKa = 7.14HH26 pKa = 6.77SPLATRR32 pKa = 11.84HH33 pKa = 5.75SPLTTHH39 pKa = 7.14HH40 pKa = 6.77SPLATRR46 pKa = 11.84HH47 pKa = 5.78SPLATRR53 pKa = 11.84HH54 pKa = 5.75SPLTTHH60 pKa = 7.14HH61 pKa = 6.76SPLTTHH67 pKa = 7.06HH68 pKa = 6.76SPLTTHH74 pKa = 7.12HH75 pKa = 6.8SPLTTRR81 pKa = 11.84YY82 pKa = 10.15SLLATHH88 pKa = 7.43RR89 pKa = 3.95
MM1 pKa = 7.23SRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84SPLAAHH11 pKa = 6.89HH12 pKa = 6.6SPLTTRR18 pKa = 11.84HH19 pKa = 5.83SPLTTHH25 pKa = 7.14HH26 pKa = 6.77SPLATRR32 pKa = 11.84HH33 pKa = 5.75SPLTTHH39 pKa = 7.14HH40 pKa = 6.77SPLATRR46 pKa = 11.84HH47 pKa = 5.78SPLATRR53 pKa = 11.84HH54 pKa = 5.75SPLTTHH60 pKa = 7.14HH61 pKa = 6.76SPLTTHH67 pKa = 7.06HH68 pKa = 6.76SPLTTHH74 pKa = 7.12HH75 pKa = 6.8SPLTTRR81 pKa = 11.84YY82 pKa = 10.15SLLATHH88 pKa = 7.43RR89 pKa = 3.95
Molecular weight: 9.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2301654 |
19 |
11909 |
411.1 |
45.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.857 ± 0.037 | 1.101 ± 0.015 |
6.649 ± 0.041 | 5.966 ± 0.027 |
3.598 ± 0.02 | 7.911 ± 0.045 |
2.178 ± 0.02 | 4.761 ± 0.021 |
2.987 ± 0.03 | 9.688 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.11 ± 0.019 | 2.916 ± 0.033 |
5.52 ± 0.028 | 4.298 ± 0.023 |
7.274 ± 0.048 | 6.632 ± 0.03 |
5.468 ± 0.037 | 7.262 ± 0.027 |
1.533 ± 0.018 | 2.291 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
