
Dysgonomonas macrotermitis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
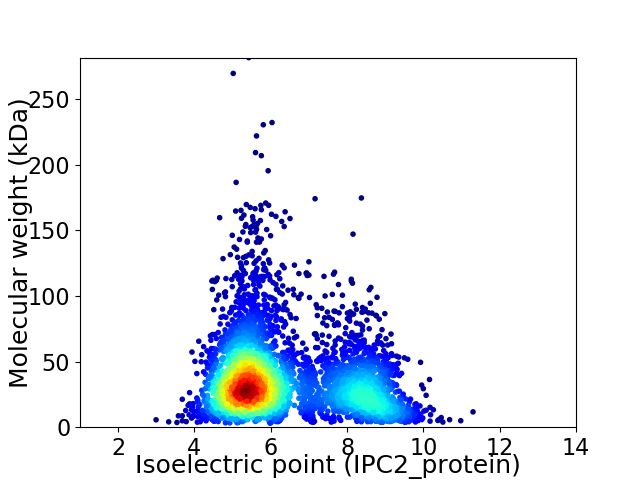
Virtual 2D-PAGE plot for 4009 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5GYG6|A0A1M5GYG6_9BACT DUF2185 domain-containing protein OS=Dysgonomonas macrotermitis OX=1346286 GN=SAMN05444362_11523 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.6SLVTTLFLSSFILFSVGCSNDD23 pKa = 3.5NNSDD27 pKa = 4.14LIDD30 pKa = 4.32GGTEE34 pKa = 3.62VTGIQQSEE42 pKa = 3.98IKK44 pKa = 10.54LRR46 pKa = 11.84IAEE49 pKa = 4.36YY50 pKa = 10.45YY51 pKa = 10.02SAPSTSAQSVSNIQTEE67 pKa = 4.25FDD69 pKa = 3.64KK70 pKa = 11.2KK71 pKa = 10.68FPNATDD77 pKa = 4.04VEE79 pKa = 4.72WKK81 pKa = 10.47ASNNVYY87 pKa = 10.53EE88 pKa = 4.38IDD90 pKa = 3.9FEE92 pKa = 4.37ISNVDD97 pKa = 3.14YY98 pKa = 10.61EE99 pKa = 4.2AWYY102 pKa = 10.13DD103 pKa = 3.89SNANLLMYY111 pKa = 10.24KK112 pKa = 10.17YY113 pKa = 10.46DD114 pKa = 3.52IANGEE119 pKa = 4.22LPSAVSSAIASDD131 pKa = 3.7YY132 pKa = 10.78TGYY135 pKa = 8.68TLDD138 pKa = 3.91EE139 pKa = 4.33AEE141 pKa = 4.46KK142 pKa = 9.87VYY144 pKa = 10.63KK145 pKa = 10.84GEE147 pKa = 4.2IIGYY151 pKa = 9.43YY152 pKa = 10.26VDD154 pKa = 3.74LKK156 pKa = 10.89KK157 pKa = 10.94NKK159 pKa = 8.75TEE161 pKa = 3.4VHH163 pKa = 6.04AFYY166 pKa = 10.91KK167 pKa = 10.63EE168 pKa = 3.96DD169 pKa = 3.29GTFISKK175 pKa = 9.94NLWEE179 pKa = 5.47DD180 pKa = 4.18DD181 pKa = 3.87SVKK184 pKa = 10.17PANDD188 pKa = 3.83AEE190 pKa = 4.53TSTPEE195 pKa = 3.7ISGTLTDD202 pKa = 4.04EE203 pKa = 4.39ATDD206 pKa = 3.87ALIAAYY212 pKa = 10.54YY213 pKa = 9.97SGNDD217 pKa = 3.4TDD219 pKa = 4.75ILPANVPSAISSNFKK234 pKa = 9.78TVFPNARR241 pKa = 11.84DD242 pKa = 3.75IDD244 pKa = 4.05WEE246 pKa = 4.4TSANVYY252 pKa = 10.4KK253 pKa = 10.73VDD255 pKa = 4.23FEE257 pKa = 5.16INNVDD262 pKa = 2.88YY263 pKa = 10.88DD264 pKa = 3.06AWYY267 pKa = 10.84NSDD270 pKa = 3.41GTLLAYY276 pKa = 10.41RR277 pKa = 11.84FDD279 pKa = 3.71ITKK282 pKa = 10.46SSLPQSVQTAISDD295 pKa = 3.66KK296 pKa = 10.64FNGYY300 pKa = 8.01TIEE303 pKa = 4.58DD304 pKa = 3.64VEE306 pKa = 4.63KK307 pKa = 10.83VIKK310 pKa = 10.59ANSAGYY316 pKa = 9.87LVEE319 pKa = 5.06LEE321 pKa = 4.13NLNIEE326 pKa = 3.78EE327 pKa = 4.24DD328 pKa = 3.92AYY330 pKa = 10.14FTEE333 pKa = 5.21DD334 pKa = 2.59GTYY337 pKa = 10.17ISNSFYY343 pKa = 10.8KK344 pKa = 10.75ASTSGDD350 pKa = 3.4NTEE353 pKa = 4.65EE354 pKa = 4.08PVTTPEE360 pKa = 3.91IPVDD364 pKa = 3.66GNYY367 pKa = 10.01TDD369 pKa = 5.81DD370 pKa = 5.59EE371 pKa = 5.0IDD373 pKa = 4.02ALLASYY379 pKa = 9.88QQGRR383 pKa = 11.84DD384 pKa = 3.12KK385 pKa = 11.01DD386 pKa = 3.58INAANVPVLVTTAFNSQFASARR408 pKa = 11.84DD409 pKa = 3.56IDD411 pKa = 3.69WDD413 pKa = 4.04YY414 pKa = 11.59VGNVYY419 pKa = 10.56KK420 pKa = 10.68VDD422 pKa = 4.07FEE424 pKa = 4.35IANVDD429 pKa = 3.4YY430 pKa = 10.29EE431 pKa = 4.12AWYY434 pKa = 10.88VSDD437 pKa = 4.21GTLLMYY443 pKa = 8.92TQEE446 pKa = 3.84VRR448 pKa = 11.84YY449 pKa = 7.59PTIASTVQNAVAAKK463 pKa = 7.44YY464 pKa = 9.05TEE466 pKa = 4.15YY467 pKa = 11.07KK468 pKa = 10.53VDD470 pKa = 3.5GCDD473 pKa = 3.35YY474 pKa = 9.91FQKK477 pKa = 10.19GTIKK481 pKa = 10.72GYY483 pKa = 10.26IIEE486 pKa = 4.73LEE488 pKa = 4.2NKK490 pKa = 8.79RR491 pKa = 11.84TDD493 pKa = 3.04AEE495 pKa = 4.53LIVIYY500 pKa = 10.64KK501 pKa = 10.02EE502 pKa = 3.95DD503 pKa = 3.51GAFISEE509 pKa = 4.08ITDD512 pKa = 3.46YY513 pKa = 11.75
MM1 pKa = 7.46KK2 pKa = 10.6SLVTTLFLSSFILFSVGCSNDD23 pKa = 3.5NNSDD27 pKa = 4.14LIDD30 pKa = 4.32GGTEE34 pKa = 3.62VTGIQQSEE42 pKa = 3.98IKK44 pKa = 10.54LRR46 pKa = 11.84IAEE49 pKa = 4.36YY50 pKa = 10.45YY51 pKa = 10.02SAPSTSAQSVSNIQTEE67 pKa = 4.25FDD69 pKa = 3.64KK70 pKa = 11.2KK71 pKa = 10.68FPNATDD77 pKa = 4.04VEE79 pKa = 4.72WKK81 pKa = 10.47ASNNVYY87 pKa = 10.53EE88 pKa = 4.38IDD90 pKa = 3.9FEE92 pKa = 4.37ISNVDD97 pKa = 3.14YY98 pKa = 10.61EE99 pKa = 4.2AWYY102 pKa = 10.13DD103 pKa = 3.89SNANLLMYY111 pKa = 10.24KK112 pKa = 10.17YY113 pKa = 10.46DD114 pKa = 3.52IANGEE119 pKa = 4.22LPSAVSSAIASDD131 pKa = 3.7YY132 pKa = 10.78TGYY135 pKa = 8.68TLDD138 pKa = 3.91EE139 pKa = 4.33AEE141 pKa = 4.46KK142 pKa = 9.87VYY144 pKa = 10.63KK145 pKa = 10.84GEE147 pKa = 4.2IIGYY151 pKa = 9.43YY152 pKa = 10.26VDD154 pKa = 3.74LKK156 pKa = 10.89KK157 pKa = 10.94NKK159 pKa = 8.75TEE161 pKa = 3.4VHH163 pKa = 6.04AFYY166 pKa = 10.91KK167 pKa = 10.63EE168 pKa = 3.96DD169 pKa = 3.29GTFISKK175 pKa = 9.94NLWEE179 pKa = 5.47DD180 pKa = 4.18DD181 pKa = 3.87SVKK184 pKa = 10.17PANDD188 pKa = 3.83AEE190 pKa = 4.53TSTPEE195 pKa = 3.7ISGTLTDD202 pKa = 4.04EE203 pKa = 4.39ATDD206 pKa = 3.87ALIAAYY212 pKa = 10.54YY213 pKa = 9.97SGNDD217 pKa = 3.4TDD219 pKa = 4.75ILPANVPSAISSNFKK234 pKa = 9.78TVFPNARR241 pKa = 11.84DD242 pKa = 3.75IDD244 pKa = 4.05WEE246 pKa = 4.4TSANVYY252 pKa = 10.4KK253 pKa = 10.73VDD255 pKa = 4.23FEE257 pKa = 5.16INNVDD262 pKa = 2.88YY263 pKa = 10.88DD264 pKa = 3.06AWYY267 pKa = 10.84NSDD270 pKa = 3.41GTLLAYY276 pKa = 10.41RR277 pKa = 11.84FDD279 pKa = 3.71ITKK282 pKa = 10.46SSLPQSVQTAISDD295 pKa = 3.66KK296 pKa = 10.64FNGYY300 pKa = 8.01TIEE303 pKa = 4.58DD304 pKa = 3.64VEE306 pKa = 4.63KK307 pKa = 10.83VIKK310 pKa = 10.59ANSAGYY316 pKa = 9.87LVEE319 pKa = 5.06LEE321 pKa = 4.13NLNIEE326 pKa = 3.78EE327 pKa = 4.24DD328 pKa = 3.92AYY330 pKa = 10.14FTEE333 pKa = 5.21DD334 pKa = 2.59GTYY337 pKa = 10.17ISNSFYY343 pKa = 10.8KK344 pKa = 10.75ASTSGDD350 pKa = 3.4NTEE353 pKa = 4.65EE354 pKa = 4.08PVTTPEE360 pKa = 3.91IPVDD364 pKa = 3.66GNYY367 pKa = 10.01TDD369 pKa = 5.81DD370 pKa = 5.59EE371 pKa = 5.0IDD373 pKa = 4.02ALLASYY379 pKa = 9.88QQGRR383 pKa = 11.84DD384 pKa = 3.12KK385 pKa = 11.01DD386 pKa = 3.58INAANVPVLVTTAFNSQFASARR408 pKa = 11.84DD409 pKa = 3.56IDD411 pKa = 3.69WDD413 pKa = 4.04YY414 pKa = 11.59VGNVYY419 pKa = 10.56KK420 pKa = 10.68VDD422 pKa = 4.07FEE424 pKa = 4.35IANVDD429 pKa = 3.4YY430 pKa = 10.29EE431 pKa = 4.12AWYY434 pKa = 10.88VSDD437 pKa = 4.21GTLLMYY443 pKa = 8.92TQEE446 pKa = 3.84VRR448 pKa = 11.84YY449 pKa = 7.59PTIASTVQNAVAAKK463 pKa = 7.44YY464 pKa = 9.05TEE466 pKa = 4.15YY467 pKa = 11.07KK468 pKa = 10.53VDD470 pKa = 3.5GCDD473 pKa = 3.35YY474 pKa = 9.91FQKK477 pKa = 10.19GTIKK481 pKa = 10.72GYY483 pKa = 10.26IIEE486 pKa = 4.73LEE488 pKa = 4.2NKK490 pKa = 8.79RR491 pKa = 11.84TDD493 pKa = 3.04AEE495 pKa = 4.53LIVIYY500 pKa = 10.64KK501 pKa = 10.02EE502 pKa = 3.95DD503 pKa = 3.51GAFISEE509 pKa = 4.08ITDD512 pKa = 3.46YY513 pKa = 11.75
Molecular weight: 57.3 kDa
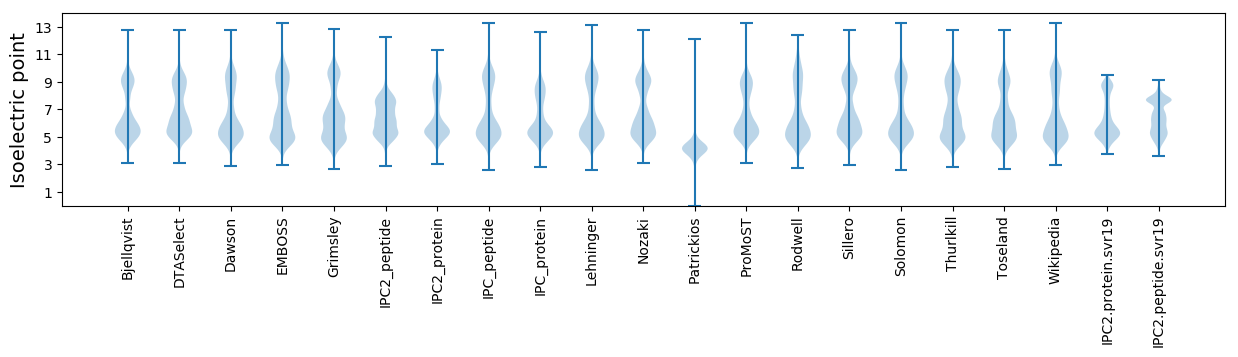
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M4X4T7|A0A1M4X4T7_9BACT TonB-linked outer membrane protein SusC/RagA family OS=Dysgonomonas macrotermitis OX=1346286 GN=SAMN05444362_102396 PE=3 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84TSRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 7.8GFRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84FRR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 6.62RR21 pKa = 11.84TRR23 pKa = 11.84FRR25 pKa = 11.84GYY27 pKa = 9.0FRR29 pKa = 11.84RR30 pKa = 11.84FRR32 pKa = 11.84SFRR35 pKa = 11.84PFRR38 pKa = 11.84RR39 pKa = 11.84FRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84SSSGRR49 pKa = 11.84GLFSGRR55 pKa = 11.84RR56 pKa = 11.84TRR58 pKa = 11.84ILGFGVPTILLVLAVIVAIWKK79 pKa = 6.33WQPIKK84 pKa = 9.83TWVSKK89 pKa = 10.19FIKK92 pKa = 10.31VV93 pKa = 3.18
MM1 pKa = 7.42ARR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84TSRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 7.8GFRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84FRR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 6.62RR21 pKa = 11.84TRR23 pKa = 11.84FRR25 pKa = 11.84GYY27 pKa = 9.0FRR29 pKa = 11.84RR30 pKa = 11.84FRR32 pKa = 11.84SFRR35 pKa = 11.84PFRR38 pKa = 11.84RR39 pKa = 11.84FRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84SSSGRR49 pKa = 11.84GLFSGRR55 pKa = 11.84RR56 pKa = 11.84TRR58 pKa = 11.84ILGFGVPTILLVLAVIVAIWKK79 pKa = 6.33WQPIKK84 pKa = 9.83TWVSKK89 pKa = 10.19FIKK92 pKa = 10.31VV93 pKa = 3.18
Molecular weight: 11.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1390717 |
28 |
2492 |
346.9 |
39.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.359 ± 0.04 | 0.955 ± 0.013 |
5.834 ± 0.026 | 6.184 ± 0.044 |
4.717 ± 0.029 | 6.584 ± 0.04 |
1.618 ± 0.018 | 7.929 ± 0.041 |
7.142 ± 0.037 | 8.945 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.016 | 5.817 ± 0.034 |
3.522 ± 0.02 | 3.358 ± 0.019 |
3.961 ± 0.025 | 6.837 ± 0.033 |
5.828 ± 0.037 | 6.133 ± 0.029 |
1.228 ± 0.016 | 4.682 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
