
Lentinula edodes negative-strand RNA virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Lentinuvirus; Lentinula lentinuvirus
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
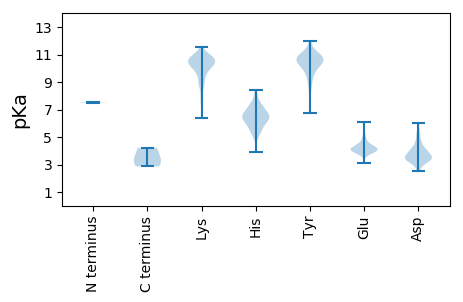
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A4P2VRN6|A0A4P2VRN6_9VIRU Replicase OS=Lentinula edodes negative-strand RNA virus 2 OX=2547431 GN=L PE=4 SV=1
MM1 pKa = 7.53NSNNKK6 pKa = 9.02LLKK9 pKa = 10.2IFGGSRR15 pKa = 11.84RR16 pKa = 11.84GKK18 pKa = 9.81VEE20 pKa = 3.7NDD22 pKa = 3.12EE23 pKa = 4.57EE24 pKa = 4.04EE25 pKa = 4.49GKK27 pKa = 10.66AVEE30 pKa = 4.48RR31 pKa = 11.84EE32 pKa = 4.03EE33 pKa = 5.3APPQAAPMKK42 pKa = 10.28HH43 pKa = 5.74FKK45 pKa = 10.51FPNKK49 pKa = 7.53YY50 pKa = 10.61ARR52 pKa = 11.84MEE54 pKa = 3.96EE55 pKa = 4.34HH56 pKa = 6.53IMNAEE61 pKa = 3.95RR62 pKa = 11.84SSGPFEE68 pKa = 4.05YY69 pKa = 10.75VAIASRR75 pKa = 11.84EE76 pKa = 3.7TSTFKK81 pKa = 11.06KK82 pKa = 10.38LLEE85 pKa = 4.32ALSVTKK91 pKa = 10.73ASVRR95 pKa = 11.84SYY97 pKa = 11.48DD98 pKa = 4.37LSFDD102 pKa = 4.14DD103 pKa = 5.33LPIYY107 pKa = 10.44NLSKK111 pKa = 10.2RR112 pKa = 11.84SSSISISSPLSLALQKK128 pKa = 10.61KK129 pKa = 9.48DD130 pKa = 5.29LHH132 pKa = 6.0FCWLKK137 pKa = 9.56EE138 pKa = 3.84VYY140 pKa = 10.12IFFVPTQSFSSSYY153 pKa = 11.25SEE155 pKa = 3.77ITFEE159 pKa = 4.95LNDD162 pKa = 3.28NRR164 pKa = 11.84FVEE167 pKa = 4.62EE168 pKa = 4.1SLVRR172 pKa = 11.84SITFPSNVGINGHH185 pKa = 6.35FSLDD189 pKa = 3.41YY190 pKa = 11.29SVFKK194 pKa = 10.73DD195 pKa = 4.14DD196 pKa = 5.32LPMISFSVKK205 pKa = 10.2CKK207 pKa = 10.5NSYY210 pKa = 10.2LKK212 pKa = 10.79EE213 pKa = 3.98GVVWGSLKK221 pKa = 10.84LVIQTKK227 pKa = 10.27FSDD230 pKa = 3.26QALIVSTTRR239 pKa = 11.84TAAILQMADD248 pKa = 3.24TTMKK252 pKa = 10.43TYY254 pKa = 10.99DD255 pKa = 4.16RR256 pKa = 11.84DD257 pKa = 4.09PNHH260 pKa = 7.37LDD262 pKa = 4.85LMFDD266 pKa = 4.09DD267 pKa = 6.02PDD269 pKa = 4.85LEE271 pKa = 4.28MMKK274 pKa = 10.46KK275 pKa = 9.77FAKK278 pKa = 9.44QGRR281 pKa = 11.84IVDD284 pKa = 3.84HH285 pKa = 6.26NQALGNRR292 pKa = 11.84EE293 pKa = 4.29RR294 pKa = 11.84IAMSGSEE301 pKa = 4.06AGSIEE306 pKa = 4.12GSQSGNSRR314 pKa = 11.84VKK316 pKa = 10.66EE317 pKa = 3.57EE318 pKa = 3.96WLRR321 pKa = 11.84GTRR324 pKa = 11.84FSRR327 pKa = 11.84GRR329 pKa = 11.84VSPVASVEE337 pKa = 3.85EE338 pKa = 4.45DD339 pKa = 3.4VRR341 pKa = 11.84SDD343 pKa = 3.54DD344 pKa = 3.99QEE346 pKa = 4.16VGEE349 pKa = 5.71FDD351 pKa = 3.64LTPNDD356 pKa = 3.8FTSSGMTEE364 pKa = 3.78RR365 pKa = 11.84AFSKK369 pKa = 10.71SAMKK373 pKa = 10.47APRR376 pKa = 11.84RR377 pKa = 11.84SLKK380 pKa = 10.77DD381 pKa = 3.18PSTSPEE387 pKa = 3.67TAANTSRR394 pKa = 11.84NEE396 pKa = 4.12GPSNWRR402 pKa = 11.84EE403 pKa = 3.91DD404 pKa = 3.67EE405 pKa = 4.19EE406 pKa = 4.4EE407 pKa = 3.96QRR409 pKa = 11.84TTARR413 pKa = 11.84NKK415 pKa = 10.01SVYY418 pKa = 9.78IYY420 pKa = 9.5EE421 pKa = 4.52GPP423 pKa = 3.37
MM1 pKa = 7.53NSNNKK6 pKa = 9.02LLKK9 pKa = 10.2IFGGSRR15 pKa = 11.84RR16 pKa = 11.84GKK18 pKa = 9.81VEE20 pKa = 3.7NDD22 pKa = 3.12EE23 pKa = 4.57EE24 pKa = 4.04EE25 pKa = 4.49GKK27 pKa = 10.66AVEE30 pKa = 4.48RR31 pKa = 11.84EE32 pKa = 4.03EE33 pKa = 5.3APPQAAPMKK42 pKa = 10.28HH43 pKa = 5.74FKK45 pKa = 10.51FPNKK49 pKa = 7.53YY50 pKa = 10.61ARR52 pKa = 11.84MEE54 pKa = 3.96EE55 pKa = 4.34HH56 pKa = 6.53IMNAEE61 pKa = 3.95RR62 pKa = 11.84SSGPFEE68 pKa = 4.05YY69 pKa = 10.75VAIASRR75 pKa = 11.84EE76 pKa = 3.7TSTFKK81 pKa = 11.06KK82 pKa = 10.38LLEE85 pKa = 4.32ALSVTKK91 pKa = 10.73ASVRR95 pKa = 11.84SYY97 pKa = 11.48DD98 pKa = 4.37LSFDD102 pKa = 4.14DD103 pKa = 5.33LPIYY107 pKa = 10.44NLSKK111 pKa = 10.2RR112 pKa = 11.84SSSISISSPLSLALQKK128 pKa = 10.61KK129 pKa = 9.48DD130 pKa = 5.29LHH132 pKa = 6.0FCWLKK137 pKa = 9.56EE138 pKa = 3.84VYY140 pKa = 10.12IFFVPTQSFSSSYY153 pKa = 11.25SEE155 pKa = 3.77ITFEE159 pKa = 4.95LNDD162 pKa = 3.28NRR164 pKa = 11.84FVEE167 pKa = 4.62EE168 pKa = 4.1SLVRR172 pKa = 11.84SITFPSNVGINGHH185 pKa = 6.35FSLDD189 pKa = 3.41YY190 pKa = 11.29SVFKK194 pKa = 10.73DD195 pKa = 4.14DD196 pKa = 5.32LPMISFSVKK205 pKa = 10.2CKK207 pKa = 10.5NSYY210 pKa = 10.2LKK212 pKa = 10.79EE213 pKa = 3.98GVVWGSLKK221 pKa = 10.84LVIQTKK227 pKa = 10.27FSDD230 pKa = 3.26QALIVSTTRR239 pKa = 11.84TAAILQMADD248 pKa = 3.24TTMKK252 pKa = 10.43TYY254 pKa = 10.99DD255 pKa = 4.16RR256 pKa = 11.84DD257 pKa = 4.09PNHH260 pKa = 7.37LDD262 pKa = 4.85LMFDD266 pKa = 4.09DD267 pKa = 6.02PDD269 pKa = 4.85LEE271 pKa = 4.28MMKK274 pKa = 10.46KK275 pKa = 9.77FAKK278 pKa = 9.44QGRR281 pKa = 11.84IVDD284 pKa = 3.84HH285 pKa = 6.26NQALGNRR292 pKa = 11.84EE293 pKa = 4.29RR294 pKa = 11.84IAMSGSEE301 pKa = 4.06AGSIEE306 pKa = 4.12GSQSGNSRR314 pKa = 11.84VKK316 pKa = 10.66EE317 pKa = 3.57EE318 pKa = 3.96WLRR321 pKa = 11.84GTRR324 pKa = 11.84FSRR327 pKa = 11.84GRR329 pKa = 11.84VSPVASVEE337 pKa = 3.85EE338 pKa = 4.45DD339 pKa = 3.4VRR341 pKa = 11.84SDD343 pKa = 3.54DD344 pKa = 3.99QEE346 pKa = 4.16VGEE349 pKa = 5.71FDD351 pKa = 3.64LTPNDD356 pKa = 3.8FTSSGMTEE364 pKa = 3.78RR365 pKa = 11.84AFSKK369 pKa = 10.71SAMKK373 pKa = 10.47APRR376 pKa = 11.84RR377 pKa = 11.84SLKK380 pKa = 10.77DD381 pKa = 3.18PSTSPEE387 pKa = 3.67TAANTSRR394 pKa = 11.84NEE396 pKa = 4.12GPSNWRR402 pKa = 11.84EE403 pKa = 3.91DD404 pKa = 3.67EE405 pKa = 4.19EE406 pKa = 4.4EE407 pKa = 3.96QRR409 pKa = 11.84TTARR413 pKa = 11.84NKK415 pKa = 10.01SVYY418 pKa = 9.78IYY420 pKa = 9.5EE421 pKa = 4.52GPP423 pKa = 3.37
Molecular weight: 47.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P2W265|A0A4P2W265_9VIRU Nucleocapsid protein OS=Lentinula edodes negative-strand RNA virus 2 OX=2547431 GN=ORF2b PE=4 SV=1
MM1 pKa = 7.5AVALRR6 pKa = 11.84NRR8 pKa = 11.84FNGLQRR14 pKa = 11.84TDD16 pKa = 2.71VSLAVGTILTQTEE29 pKa = 3.8ISAVLEE35 pKa = 3.74ADD37 pKa = 3.02INIIQEE43 pKa = 4.4RR44 pKa = 11.84IAGEE48 pKa = 3.89FIIDD52 pKa = 3.43VDD54 pKa = 4.66SINAALEE61 pKa = 3.95LLSYY65 pKa = 10.79EE66 pKa = 4.27GFDD69 pKa = 3.45PRR71 pKa = 11.84IVYY74 pKa = 10.15AHH76 pKa = 7.49LAHH79 pKa = 6.45IQKK82 pKa = 10.12AGNIGWPQLKK92 pKa = 9.52EE93 pKa = 4.02DD94 pKa = 4.18LMIMLVIHH102 pKa = 6.29HH103 pKa = 6.36VRR105 pKa = 11.84GNINSNNMKK114 pKa = 10.37SMKK117 pKa = 10.25SEE119 pKa = 3.77AKK121 pKa = 10.54NLINEE126 pKa = 4.75LFAKK130 pKa = 8.94YY131 pKa = 9.97HH132 pKa = 5.65IVVAKK137 pKa = 8.73DD138 pKa = 3.06TSKK141 pKa = 11.12RR142 pKa = 11.84LAVTLPRR149 pKa = 11.84LGASFPYY156 pKa = 10.08QVAQIAKK163 pKa = 8.66ICNRR167 pKa = 11.84DD168 pKa = 3.11FAGFCEE174 pKa = 4.62SAVLPAAMKK183 pKa = 9.13STSFPSLVPKK193 pKa = 9.02STKK196 pKa = 9.63ISLLMTVAYY205 pKa = 9.79NAYY208 pKa = 9.08ATDD211 pKa = 3.49QACALKK217 pKa = 10.39KK218 pKa = 10.59LNYY221 pKa = 10.31LEE223 pKa = 4.38MKK225 pKa = 10.61EE226 pKa = 4.0EE227 pKa = 3.84DD228 pKa = 3.39RR229 pKa = 11.84KK230 pKa = 10.82RR231 pKa = 11.84LFLEE235 pKa = 3.68QQRR238 pKa = 11.84YY239 pKa = 6.76TDD241 pKa = 3.06ISYY244 pKa = 10.94NSPVLDD250 pKa = 3.72KK251 pKa = 11.11DD252 pKa = 3.42HH253 pKa = 6.87RR254 pKa = 11.84QAALFNLGLGNSTVYY269 pKa = 10.48KK270 pKa = 10.76ALLSCTKK277 pKa = 9.83YY278 pKa = 10.99SSVDD282 pKa = 3.07TSLFCSEE289 pKa = 4.2TEE291 pKa = 3.94YY292 pKa = 11.1LGEE295 pKa = 4.51IISYY299 pKa = 9.23QASADD304 pKa = 4.34LIAKK308 pKa = 7.94MVASRR313 pKa = 11.84VSGPSS318 pKa = 2.87
MM1 pKa = 7.5AVALRR6 pKa = 11.84NRR8 pKa = 11.84FNGLQRR14 pKa = 11.84TDD16 pKa = 2.71VSLAVGTILTQTEE29 pKa = 3.8ISAVLEE35 pKa = 3.74ADD37 pKa = 3.02INIIQEE43 pKa = 4.4RR44 pKa = 11.84IAGEE48 pKa = 3.89FIIDD52 pKa = 3.43VDD54 pKa = 4.66SINAALEE61 pKa = 3.95LLSYY65 pKa = 10.79EE66 pKa = 4.27GFDD69 pKa = 3.45PRR71 pKa = 11.84IVYY74 pKa = 10.15AHH76 pKa = 7.49LAHH79 pKa = 6.45IQKK82 pKa = 10.12AGNIGWPQLKK92 pKa = 9.52EE93 pKa = 4.02DD94 pKa = 4.18LMIMLVIHH102 pKa = 6.29HH103 pKa = 6.36VRR105 pKa = 11.84GNINSNNMKK114 pKa = 10.37SMKK117 pKa = 10.25SEE119 pKa = 3.77AKK121 pKa = 10.54NLINEE126 pKa = 4.75LFAKK130 pKa = 8.94YY131 pKa = 9.97HH132 pKa = 5.65IVVAKK137 pKa = 8.73DD138 pKa = 3.06TSKK141 pKa = 11.12RR142 pKa = 11.84LAVTLPRR149 pKa = 11.84LGASFPYY156 pKa = 10.08QVAQIAKK163 pKa = 8.66ICNRR167 pKa = 11.84DD168 pKa = 3.11FAGFCEE174 pKa = 4.62SAVLPAAMKK183 pKa = 9.13STSFPSLVPKK193 pKa = 9.02STKK196 pKa = 9.63ISLLMTVAYY205 pKa = 9.79NAYY208 pKa = 9.08ATDD211 pKa = 3.49QACALKK217 pKa = 10.39KK218 pKa = 10.59LNYY221 pKa = 10.31LEE223 pKa = 4.38MKK225 pKa = 10.61EE226 pKa = 4.0EE227 pKa = 3.84DD228 pKa = 3.39RR229 pKa = 11.84KK230 pKa = 10.82RR231 pKa = 11.84LFLEE235 pKa = 3.68QQRR238 pKa = 11.84YY239 pKa = 6.76TDD241 pKa = 3.06ISYY244 pKa = 10.94NSPVLDD250 pKa = 3.72KK251 pKa = 11.11DD252 pKa = 3.42HH253 pKa = 6.87RR254 pKa = 11.84QAALFNLGLGNSTVYY269 pKa = 10.48KK270 pKa = 10.76ALLSCTKK277 pKa = 9.83YY278 pKa = 10.99SSVDD282 pKa = 3.07TSLFCSEE289 pKa = 4.2TEE291 pKa = 3.94YY292 pKa = 11.1LGEE295 pKa = 4.51IISYY299 pKa = 9.23QASADD304 pKa = 4.34LIAKK308 pKa = 7.94MVASRR313 pKa = 11.84VSGPSS318 pKa = 2.87
Molecular weight: 35.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3050 |
318 |
2309 |
1016.7 |
116.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.656 ± 1.937 | 1.377 ± 0.285 |
6.197 ± 0.459 | 8.23 ± 0.746 |
5.344 ± 0.511 | 4.492 ± 0.152 |
2.328 ± 0.373 | 6.361 ± 0.644 |
7.738 ± 0.59 | 8.918 ± 0.695 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.115 ± 0.086 | 4.852 ± 0.156 |
3.639 ± 0.281 | 2.492 ± 0.373 |
5.508 ± 0.372 | 9.508 ± 1.028 |
4.918 ± 0.088 | 5.902 ± 0.103 |
1.049 ± 0.221 | 3.377 ± 0.254 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
