
Tolypocladium paradoxum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Ophiocordycipitaceae; Tolypocladium
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
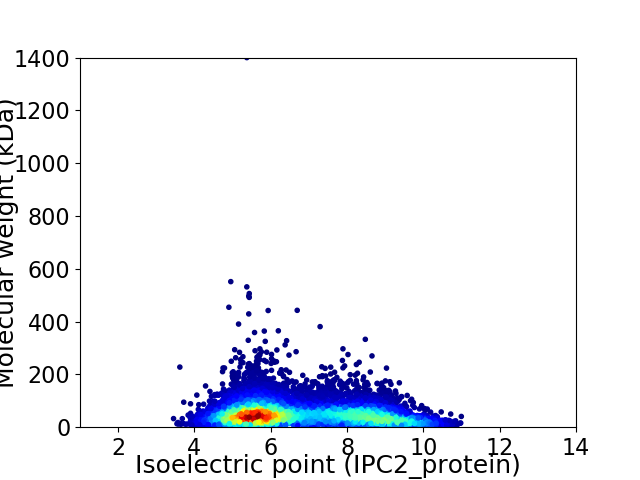
Virtual 2D-PAGE plot for 8979 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S4KU88|A0A2S4KU88_9HYPO Matrin-type domain-containing protein OS=Tolypocladium paradoxum OX=94208 GN=TPAR_06016 PE=3 SV=1
MM1 pKa = 6.73STKK4 pKa = 9.85ILYY7 pKa = 9.86DD8 pKa = 3.43AVVGPGANEE17 pKa = 3.91WYY19 pKa = 7.75GTLVVTNIRR28 pKa = 11.84YY29 pKa = 9.92DD30 pKa = 3.21GGGTVTVQKK39 pKa = 10.63FLGVEE44 pKa = 4.34FLSPATVAATDD55 pKa = 3.74INVLTNPWVQVTPEE69 pKa = 4.0VTNEE73 pKa = 4.08QIDD76 pKa = 3.73SSTFSITAKK85 pKa = 10.58LNVDD89 pKa = 3.37NSVSLTGTTITIGINGDD106 pKa = 3.48LTKK109 pKa = 11.06DD110 pKa = 3.0SDD112 pKa = 3.9RR113 pKa = 11.84YY114 pKa = 6.25TTSFVFVADD123 pKa = 5.43GIPDD127 pKa = 3.41TSGTVNVDD135 pKa = 3.7CAASPDD141 pKa = 3.8PALDD145 pKa = 4.0GEE147 pKa = 4.48QQAVTFTLGGRR158 pKa = 11.84VVNVTVPLGEE168 pKa = 4.1TTPAKK173 pKa = 10.57VPAGTYY179 pKa = 10.44DD180 pKa = 3.22VTAAEE185 pKa = 4.8LTTDD189 pKa = 3.18AQTVVASAQVSPTSVTVVAGEE210 pKa = 4.27TVDD213 pKa = 3.9VAVTYY218 pKa = 10.85DD219 pKa = 3.62LVEE222 pKa = 4.56KK223 pKa = 10.84YY224 pKa = 10.55SAIDD228 pKa = 3.48VTIGTILPLEE238 pKa = 4.44KK239 pKa = 10.45EE240 pKa = 4.15QFHH243 pKa = 5.72VQVVDD248 pKa = 3.56QGSGTILADD257 pKa = 4.29FSSPINQTTSLRR269 pKa = 11.84RR270 pKa = 11.84LPASGTAVVSVDD282 pKa = 2.9GTTLNNVEE290 pKa = 3.87YY291 pKa = 10.39SFAVKK296 pKa = 10.51SEE298 pKa = 4.2DD299 pKa = 3.58LSPSLFQVSFSQDD312 pKa = 3.21DD313 pKa = 4.0VTTTPVDD320 pKa = 3.27TTGFVTLPIVVGTDD334 pKa = 2.69ITLDD338 pKa = 3.23ATISVRR344 pKa = 11.84LTSSTNYY351 pKa = 10.14VYY353 pKa = 10.09TQDD356 pKa = 3.28VKK358 pKa = 11.22AAAGATNFAVPVGPGYY374 pKa = 9.36YY375 pKa = 8.77TVQAASIINNGIVYY389 pKa = 9.28VVSISATSLTVKK401 pKa = 10.57ADD403 pKa = 3.44GSSTVQLTLQRR414 pKa = 11.84GANLNVRR421 pKa = 11.84GFPGFLSFGGCADD434 pKa = 3.74LTSGNQADD442 pKa = 4.51FAAALASSVFKK453 pKa = 11.25YY454 pKa = 10.58AGTDD458 pKa = 3.5GAGDD462 pKa = 3.71AGSFLTDD469 pKa = 3.52DD470 pKa = 3.17QATRR474 pKa = 11.84QTIALARR481 pKa = 11.84AVEE484 pKa = 4.21TQVADD489 pKa = 4.08GNPVLPVMISYY500 pKa = 7.82TCNMSGGNPADD511 pKa = 3.56HH512 pKa = 6.54LQNVDD517 pKa = 3.35GLAHH521 pKa = 6.57SFANLILSMNIATATIDD538 pKa = 3.68DD539 pKa = 4.11QHH541 pKa = 6.7PVPAGYY547 pKa = 9.93VVNPDD552 pKa = 4.79FIGACQQDD560 pKa = 4.5GLSADD565 pKa = 3.45YY566 pKa = 11.3AMQVRR571 pKa = 11.84APLTTALDD579 pKa = 3.49HH580 pKa = 6.59WSVSAEE586 pKa = 3.8IPSTITEE593 pKa = 4.17DD594 pKa = 3.19LRR596 pKa = 11.84GYY598 pKa = 9.93VLAVNWLIRR607 pKa = 11.84TVAPAVTFGWQVNLWGVGASTWVYY631 pKa = 11.01QDD633 pKa = 5.21DD634 pKa = 4.27DD635 pKa = 3.79VADD638 pKa = 3.87IARR641 pKa = 11.84QTGDD645 pKa = 3.23YY646 pKa = 10.76AKK648 pKa = 9.85TLGVFDD654 pKa = 5.26GDD656 pKa = 3.83NLPDD660 pKa = 4.05FLAVDD665 pKa = 4.68RR666 pKa = 11.84YY667 pKa = 10.05EE668 pKa = 5.88ADD670 pKa = 3.55DD671 pKa = 3.82FTVRR675 pKa = 11.84AYY677 pKa = 10.91GNGYY681 pKa = 10.25CYY683 pKa = 10.58GPRR686 pKa = 11.84EE687 pKa = 3.94WGRR690 pKa = 11.84FFDD693 pKa = 4.58FCAALSAQLQFPIMPWQIPSSWTPLTTDD721 pKa = 3.21AVNDD725 pKa = 3.86NFDD728 pKa = 3.65SQHH731 pKa = 6.01WGTAGSYY738 pKa = 10.86IMGDD742 pKa = 3.33PGIGSDD748 pKa = 3.68YY749 pKa = 11.11HH750 pKa = 6.62NVNPKK755 pKa = 9.64ILALQFSPALPYY767 pKa = 9.87MGSTAEE773 pKa = 4.81DD774 pKa = 3.33IFTRR778 pKa = 11.84AEE780 pKa = 4.14PFDD783 pKa = 4.24LTSPAYY789 pKa = 10.6SDD791 pKa = 3.71FPLRR795 pKa = 11.84GIFTVLLGGGSTTGIVSSIGNPQPWVRR822 pKa = 11.84DD823 pKa = 3.33KK824 pKa = 11.41LYY826 pKa = 10.49TYY828 pKa = 7.47MQNSISFNDD837 pKa = 3.36
MM1 pKa = 6.73STKK4 pKa = 9.85ILYY7 pKa = 9.86DD8 pKa = 3.43AVVGPGANEE17 pKa = 3.91WYY19 pKa = 7.75GTLVVTNIRR28 pKa = 11.84YY29 pKa = 9.92DD30 pKa = 3.21GGGTVTVQKK39 pKa = 10.63FLGVEE44 pKa = 4.34FLSPATVAATDD55 pKa = 3.74INVLTNPWVQVTPEE69 pKa = 4.0VTNEE73 pKa = 4.08QIDD76 pKa = 3.73SSTFSITAKK85 pKa = 10.58LNVDD89 pKa = 3.37NSVSLTGTTITIGINGDD106 pKa = 3.48LTKK109 pKa = 11.06DD110 pKa = 3.0SDD112 pKa = 3.9RR113 pKa = 11.84YY114 pKa = 6.25TTSFVFVADD123 pKa = 5.43GIPDD127 pKa = 3.41TSGTVNVDD135 pKa = 3.7CAASPDD141 pKa = 3.8PALDD145 pKa = 4.0GEE147 pKa = 4.48QQAVTFTLGGRR158 pKa = 11.84VVNVTVPLGEE168 pKa = 4.1TTPAKK173 pKa = 10.57VPAGTYY179 pKa = 10.44DD180 pKa = 3.22VTAAEE185 pKa = 4.8LTTDD189 pKa = 3.18AQTVVASAQVSPTSVTVVAGEE210 pKa = 4.27TVDD213 pKa = 3.9VAVTYY218 pKa = 10.85DD219 pKa = 3.62LVEE222 pKa = 4.56KK223 pKa = 10.84YY224 pKa = 10.55SAIDD228 pKa = 3.48VTIGTILPLEE238 pKa = 4.44KK239 pKa = 10.45EE240 pKa = 4.15QFHH243 pKa = 5.72VQVVDD248 pKa = 3.56QGSGTILADD257 pKa = 4.29FSSPINQTTSLRR269 pKa = 11.84RR270 pKa = 11.84LPASGTAVVSVDD282 pKa = 2.9GTTLNNVEE290 pKa = 3.87YY291 pKa = 10.39SFAVKK296 pKa = 10.51SEE298 pKa = 4.2DD299 pKa = 3.58LSPSLFQVSFSQDD312 pKa = 3.21DD313 pKa = 4.0VTTTPVDD320 pKa = 3.27TTGFVTLPIVVGTDD334 pKa = 2.69ITLDD338 pKa = 3.23ATISVRR344 pKa = 11.84LTSSTNYY351 pKa = 10.14VYY353 pKa = 10.09TQDD356 pKa = 3.28VKK358 pKa = 11.22AAAGATNFAVPVGPGYY374 pKa = 9.36YY375 pKa = 8.77TVQAASIINNGIVYY389 pKa = 9.28VVSISATSLTVKK401 pKa = 10.57ADD403 pKa = 3.44GSSTVQLTLQRR414 pKa = 11.84GANLNVRR421 pKa = 11.84GFPGFLSFGGCADD434 pKa = 3.74LTSGNQADD442 pKa = 4.51FAAALASSVFKK453 pKa = 11.25YY454 pKa = 10.58AGTDD458 pKa = 3.5GAGDD462 pKa = 3.71AGSFLTDD469 pKa = 3.52DD470 pKa = 3.17QATRR474 pKa = 11.84QTIALARR481 pKa = 11.84AVEE484 pKa = 4.21TQVADD489 pKa = 4.08GNPVLPVMISYY500 pKa = 7.82TCNMSGGNPADD511 pKa = 3.56HH512 pKa = 6.54LQNVDD517 pKa = 3.35GLAHH521 pKa = 6.57SFANLILSMNIATATIDD538 pKa = 3.68DD539 pKa = 4.11QHH541 pKa = 6.7PVPAGYY547 pKa = 9.93VVNPDD552 pKa = 4.79FIGACQQDD560 pKa = 4.5GLSADD565 pKa = 3.45YY566 pKa = 11.3AMQVRR571 pKa = 11.84APLTTALDD579 pKa = 3.49HH580 pKa = 6.59WSVSAEE586 pKa = 3.8IPSTITEE593 pKa = 4.17DD594 pKa = 3.19LRR596 pKa = 11.84GYY598 pKa = 9.93VLAVNWLIRR607 pKa = 11.84TVAPAVTFGWQVNLWGVGASTWVYY631 pKa = 11.01QDD633 pKa = 5.21DD634 pKa = 4.27DD635 pKa = 3.79VADD638 pKa = 3.87IARR641 pKa = 11.84QTGDD645 pKa = 3.23YY646 pKa = 10.76AKK648 pKa = 9.85TLGVFDD654 pKa = 5.26GDD656 pKa = 3.83NLPDD660 pKa = 4.05FLAVDD665 pKa = 4.68RR666 pKa = 11.84YY667 pKa = 10.05EE668 pKa = 5.88ADD670 pKa = 3.55DD671 pKa = 3.82FTVRR675 pKa = 11.84AYY677 pKa = 10.91GNGYY681 pKa = 10.25CYY683 pKa = 10.58GPRR686 pKa = 11.84EE687 pKa = 3.94WGRR690 pKa = 11.84FFDD693 pKa = 4.58FCAALSAQLQFPIMPWQIPSSWTPLTTDD721 pKa = 3.21AVNDD725 pKa = 3.86NFDD728 pKa = 3.65SQHH731 pKa = 6.01WGTAGSYY738 pKa = 10.86IMGDD742 pKa = 3.33PGIGSDD748 pKa = 3.68YY749 pKa = 11.11HH750 pKa = 6.62NVNPKK755 pKa = 9.64ILALQFSPALPYY767 pKa = 9.87MGSTAEE773 pKa = 4.81DD774 pKa = 3.33IFTRR778 pKa = 11.84AEE780 pKa = 4.14PFDD783 pKa = 4.24LTSPAYY789 pKa = 10.6SDD791 pKa = 3.71FPLRR795 pKa = 11.84GIFTVLLGGGSTTGIVSSIGNPQPWVRR822 pKa = 11.84DD823 pKa = 3.33KK824 pKa = 11.41LYY826 pKa = 10.49TYY828 pKa = 7.47MQNSISFNDD837 pKa = 3.36
Molecular weight: 88.62 kDa
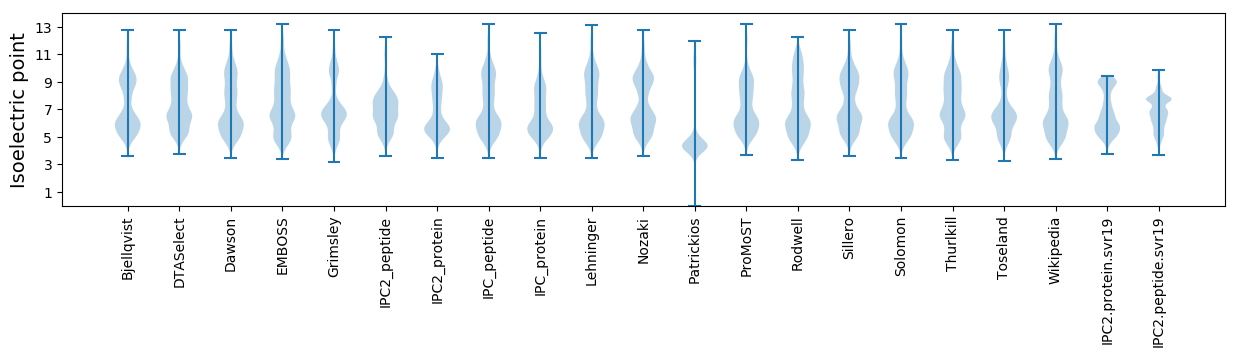
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S4L963|A0A2S4L963_9HYPO Uncharacterized protein (Fragment) OS=Tolypocladium paradoxum OX=94208 GN=TPAR_00843 PE=4 SV=1
PP1 pKa = 7.28LGRR4 pKa = 11.84ARR6 pKa = 11.84RR7 pKa = 11.84HH8 pKa = 3.97QRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84ALRR15 pKa = 11.84PEE17 pKa = 4.07PQPGRR22 pKa = 11.84RR23 pKa = 11.84LPRR26 pKa = 11.84VRR28 pKa = 11.84HH29 pKa = 5.65HH30 pKa = 6.87RR31 pKa = 11.84RR32 pKa = 11.84AAPARR37 pKa = 11.84PLRR40 pKa = 11.84GAAPRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84PHH50 pKa = 5.27RR51 pKa = 11.84QPRR54 pKa = 11.84RR55 pKa = 11.84QRR57 pKa = 11.84RR58 pKa = 11.84EE59 pKa = 3.09RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84PRR64 pKa = 11.84RR65 pKa = 11.84LQARR69 pKa = 11.84QRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84HH74 pKa = 4.56RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84LPGHH84 pKa = 6.55RR85 pKa = 11.84RR86 pKa = 11.84QGPRR90 pKa = 11.84VHH92 pKa = 6.71EE93 pKa = 4.41LGEE96 pKa = 4.02RR97 pKa = 11.84RR98 pKa = 11.84QRR100 pKa = 11.84QPDD103 pKa = 3.71GPAEE107 pKa = 4.19CRR109 pKa = 11.84RR110 pKa = 11.84AVAGRR115 pKa = 11.84SRR117 pKa = 11.84GAAARR122 pKa = 11.84VLLGDD127 pKa = 3.24QGRR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84AARR135 pKa = 11.84PRR137 pKa = 11.84RR138 pKa = 11.84QPPVPLQPPHH148 pKa = 6.66HH149 pKa = 6.79RR150 pKa = 11.84PPPRR154 pKa = 11.84RR155 pKa = 11.84HH156 pKa = 5.43LRR158 pKa = 11.84RR159 pKa = 11.84GRR161 pKa = 11.84RR162 pKa = 11.84PGPRR166 pKa = 11.84RR167 pKa = 11.84PAARR171 pKa = 11.84PPLQPDD177 pKa = 2.97QRR179 pKa = 11.84PARR182 pKa = 11.84RR183 pKa = 11.84QPQHH187 pKa = 6.31LRR189 pKa = 11.84LHH191 pKa = 6.83LRR193 pKa = 11.84RR194 pKa = 11.84QRR196 pKa = 11.84RR197 pKa = 11.84PRR199 pKa = 11.84PPPRR203 pKa = 11.84RR204 pKa = 11.84PRR206 pKa = 11.84PPRR209 pKa = 11.84RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84LRR215 pKa = 11.84PAPRR219 pKa = 11.84PPKK222 pKa = 9.63GRR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84ALLRR230 pKa = 11.84HH231 pKa = 5.92KK232 pKa = 10.47RR233 pKa = 11.84RR234 pKa = 11.84ALLLLGLCPRR244 pKa = 11.84RR245 pKa = 11.84LARR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84PRR252 pKa = 11.84ARR254 pKa = 11.84HH255 pKa = 5.29QGRR258 pKa = 11.84LDD260 pKa = 3.51PPPRR264 pKa = 11.84PRR266 pKa = 11.84HH267 pKa = 4.73LHH269 pKa = 5.75GLLQRR274 pKa = 11.84GLQQHH279 pKa = 6.29HH280 pKa = 6.19RR281 pKa = 11.84QAPHH285 pKa = 7.28LYY287 pKa = 9.28PPARR291 pKa = 11.84RR292 pKa = 11.84HH293 pKa = 4.66GHH295 pKa = 6.31HH296 pKa = 7.47DD297 pKa = 3.46ALLQHH302 pKa = 6.53HH303 pKa = 6.5QGQEE307 pKa = 3.89RR308 pKa = 11.84PALRR312 pKa = 11.84AAVDD316 pKa = 3.7PAGGRR321 pKa = 11.84HH322 pKa = 4.92PRR324 pKa = 11.84RR325 pKa = 11.84RR326 pKa = 11.84LVPGAGEE333 pKa = 3.99QGPHH337 pKa = 5.46QGAII341 pKa = 3.55
PP1 pKa = 7.28LGRR4 pKa = 11.84ARR6 pKa = 11.84RR7 pKa = 11.84HH8 pKa = 3.97QRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84ALRR15 pKa = 11.84PEE17 pKa = 4.07PQPGRR22 pKa = 11.84RR23 pKa = 11.84LPRR26 pKa = 11.84VRR28 pKa = 11.84HH29 pKa = 5.65HH30 pKa = 6.87RR31 pKa = 11.84RR32 pKa = 11.84AAPARR37 pKa = 11.84PLRR40 pKa = 11.84GAAPRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84PHH50 pKa = 5.27RR51 pKa = 11.84QPRR54 pKa = 11.84RR55 pKa = 11.84QRR57 pKa = 11.84RR58 pKa = 11.84EE59 pKa = 3.09RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84PRR64 pKa = 11.84RR65 pKa = 11.84LQARR69 pKa = 11.84QRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84HH74 pKa = 4.56RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84LPGHH84 pKa = 6.55RR85 pKa = 11.84RR86 pKa = 11.84QGPRR90 pKa = 11.84VHH92 pKa = 6.71EE93 pKa = 4.41LGEE96 pKa = 4.02RR97 pKa = 11.84RR98 pKa = 11.84QRR100 pKa = 11.84QPDD103 pKa = 3.71GPAEE107 pKa = 4.19CRR109 pKa = 11.84RR110 pKa = 11.84AVAGRR115 pKa = 11.84SRR117 pKa = 11.84GAAARR122 pKa = 11.84VLLGDD127 pKa = 3.24QGRR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84AARR135 pKa = 11.84PRR137 pKa = 11.84RR138 pKa = 11.84QPPVPLQPPHH148 pKa = 6.66HH149 pKa = 6.79RR150 pKa = 11.84PPPRR154 pKa = 11.84RR155 pKa = 11.84HH156 pKa = 5.43LRR158 pKa = 11.84RR159 pKa = 11.84GRR161 pKa = 11.84RR162 pKa = 11.84PGPRR166 pKa = 11.84RR167 pKa = 11.84PAARR171 pKa = 11.84PPLQPDD177 pKa = 2.97QRR179 pKa = 11.84PARR182 pKa = 11.84RR183 pKa = 11.84QPQHH187 pKa = 6.31LRR189 pKa = 11.84LHH191 pKa = 6.83LRR193 pKa = 11.84RR194 pKa = 11.84QRR196 pKa = 11.84RR197 pKa = 11.84PRR199 pKa = 11.84PPPRR203 pKa = 11.84RR204 pKa = 11.84PRR206 pKa = 11.84PPRR209 pKa = 11.84RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84LRR215 pKa = 11.84PAPRR219 pKa = 11.84PPKK222 pKa = 9.63GRR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84ALLRR230 pKa = 11.84HH231 pKa = 5.92KK232 pKa = 10.47RR233 pKa = 11.84RR234 pKa = 11.84ALLLLGLCPRR244 pKa = 11.84RR245 pKa = 11.84LARR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84PRR252 pKa = 11.84ARR254 pKa = 11.84HH255 pKa = 5.29QGRR258 pKa = 11.84LDD260 pKa = 3.51PPPRR264 pKa = 11.84PRR266 pKa = 11.84HH267 pKa = 4.73LHH269 pKa = 5.75GLLQRR274 pKa = 11.84GLQQHH279 pKa = 6.29HH280 pKa = 6.19RR281 pKa = 11.84QAPHH285 pKa = 7.28LYY287 pKa = 9.28PPARR291 pKa = 11.84RR292 pKa = 11.84HH293 pKa = 4.66GHH295 pKa = 6.31HH296 pKa = 7.47DD297 pKa = 3.46ALLQHH302 pKa = 6.53HH303 pKa = 6.5QGQEE307 pKa = 3.89RR308 pKa = 11.84PALRR312 pKa = 11.84AAVDD316 pKa = 3.7PAGGRR321 pKa = 11.84HH322 pKa = 4.92PRR324 pKa = 11.84RR325 pKa = 11.84RR326 pKa = 11.84LVPGAGEE333 pKa = 3.99QGPHH337 pKa = 5.46QGAII341 pKa = 3.55
Molecular weight: 40.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4469941 |
8 |
12706 |
497.8 |
54.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.653 ± 0.025 | 1.29 ± 0.008 |
5.913 ± 0.02 | 5.879 ± 0.025 |
3.512 ± 0.015 | 7.269 ± 0.023 |
2.481 ± 0.012 | 4.208 ± 0.018 |
4.487 ± 0.022 | 8.828 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.259 ± 0.01 | 3.257 ± 0.013 |
6.364 ± 0.026 | 3.998 ± 0.017 |
6.816 ± 0.025 | 7.958 ± 0.026 |
5.587 ± 0.017 | 6.293 ± 0.022 |
1.434 ± 0.009 | 2.503 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
