
Serinicoccus hydrothermalis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Ornithinimicrobiaceae; Serinicoccus
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
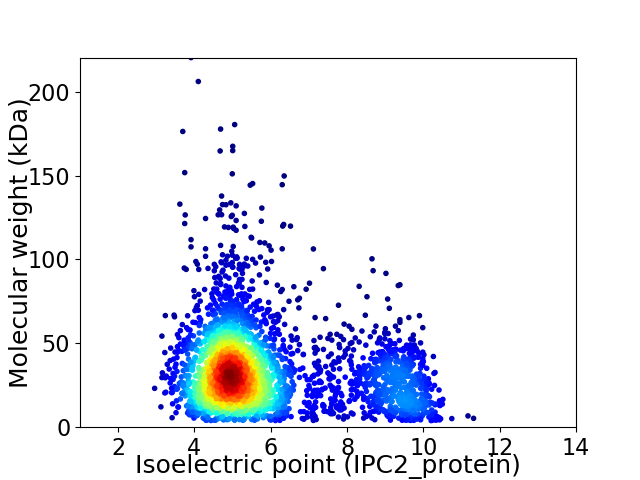
Virtual 2D-PAGE plot for 3300 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B1NAZ1|A0A1B1NAZ1_9MICO N-acetylmuramoyl-L-alanine amidase OS=Serinicoccus hydrothermalis OX=1758689 GN=SGUI_1196 PE=3 SV=1
MM1 pKa = 7.12HH2 pKa = 6.79QPSARR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84TRR11 pKa = 11.84LLALAGAGAVIAPLALVAPSAWANPDD37 pKa = 3.16LTVNEE42 pKa = 4.7DD43 pKa = 2.74GTYY46 pKa = 10.44IVMLADD52 pKa = 3.7SSLAAYY58 pKa = 10.12DD59 pKa = 4.07GGVQGIPATKK69 pKa = 9.11PAEE72 pKa = 4.25GEE74 pKa = 4.19KK75 pKa = 10.88LSTTTAAAEE84 pKa = 4.59DD85 pKa = 3.73YY86 pKa = 8.93TAYY89 pKa = 11.08LDD91 pKa = 3.79RR92 pKa = 11.84QQNAVLDD99 pKa = 4.21SVGLDD104 pKa = 3.18RR105 pKa = 11.84ADD107 pKa = 3.5AVYY110 pKa = 9.22TYY112 pKa = 9.72STAFNGFTADD122 pKa = 3.48LTGSQVSALRR132 pKa = 11.84KK133 pKa = 8.91DD134 pKa = 3.78PKK136 pKa = 10.71VAAVWEE142 pKa = 4.24NEE144 pKa = 3.44VRR146 pKa = 11.84YY147 pKa = 8.16TDD149 pKa = 4.23TITTPDD155 pKa = 3.76YY156 pKa = 11.38LGLTGDD162 pKa = 3.78DD163 pKa = 4.36GVWNTQFGGDD173 pKa = 3.63ANAGSGIVVGIIDD186 pKa = 3.49TGFWPEE192 pKa = 4.06NPSFAALPGDD202 pKa = 4.35PEE204 pKa = 5.42PPADD208 pKa = 3.49WNGEE212 pKa = 4.25CVEE215 pKa = 4.84GDD217 pKa = 3.74SADD220 pKa = 3.63PADD223 pKa = 4.68NVTCNSKK230 pKa = 10.85VIGARR235 pKa = 11.84FYY237 pKa = 10.81PDD239 pKa = 3.25TNDD242 pKa = 2.93TSFDD246 pKa = 3.75FEE248 pKa = 4.87SPRR251 pKa = 11.84DD252 pKa = 4.0TNGHH256 pKa = 5.84GSHH259 pKa = 6.62VGGTSAGNVDD269 pKa = 3.8VPMEE273 pKa = 4.3INGVEE278 pKa = 4.02LGEE281 pKa = 4.37GSGMAPAAHH290 pKa = 6.54LAFYY294 pKa = 9.93KK295 pKa = 10.76ALWQTEE301 pKa = 4.16DD302 pKa = 3.76GNGSGTSAGLVAAIDD317 pKa = 3.86DD318 pKa = 4.13AVADD322 pKa = 4.22GVDD325 pKa = 3.67VINYY329 pKa = 8.3SVSGSSDD336 pKa = 3.52YY337 pKa = 11.53VVTADD342 pKa = 4.17EE343 pKa = 5.11LAFLDD348 pKa = 4.1AANAGVFVSTSAGNSGDD365 pKa = 3.76TVGEE369 pKa = 3.8SSVAHH374 pKa = 5.73NSPWTMTVAASTHH387 pKa = 6.34DD388 pKa = 3.59RR389 pKa = 11.84TNSAEE394 pKa = 3.99VEE396 pKa = 4.3LGGGAPVTRR405 pKa = 11.84ISGTNRR411 pKa = 11.84YY412 pKa = 7.67DD413 pKa = 3.11TAAQIAAAYY422 pKa = 8.52PEE424 pKa = 4.84GVDD427 pKa = 3.21TVYY430 pKa = 10.64IATGNQFADD439 pKa = 3.78ALSGAAAASQGLVPANGLDD458 pKa = 3.77QPAVAPDD465 pKa = 4.26GSPAPVLLTKK475 pKa = 10.35VDD477 pKa = 4.95EE478 pKa = 4.76IPSATMEE485 pKa = 4.38ALEE488 pKa = 5.81SIDD491 pKa = 3.56PDD493 pKa = 3.88NIVVLGGEE501 pKa = 4.41VAVSDD506 pKa = 4.59DD507 pKa = 4.02VEE509 pKa = 4.7DD510 pKa = 4.14ALGDD514 pKa = 3.73YY515 pKa = 8.54GTVTRR520 pKa = 11.84VAGANRR526 pKa = 11.84YY527 pKa = 6.69EE528 pKa = 4.3TAALVAEE535 pKa = 4.58QYY537 pKa = 11.53GEE539 pKa = 3.93VDD541 pKa = 3.17HH542 pKa = 7.02VYY544 pKa = 10.61VATGQDD550 pKa = 3.19EE551 pKa = 4.73AFPDD555 pKa = 3.94ALSGSALAGSEE566 pKa = 4.19GVPVLLTKK574 pKa = 10.65SDD576 pKa = 3.88MVPGAVSDD584 pKa = 3.96ALEE587 pKa = 4.21AMGDD591 pKa = 3.87PEE593 pKa = 4.19VHH595 pKa = 6.62VIGGSAAVSEE605 pKa = 5.23DD606 pKa = 3.26VFEE609 pKa = 5.02DD610 pKa = 3.31LGGSEE615 pKa = 4.52RR616 pKa = 11.84LSGTDD621 pKa = 2.83RR622 pKa = 11.84YY623 pKa = 8.48GTSVAVAEE631 pKa = 4.25QFGYY635 pKa = 10.81DD636 pKa = 3.81ADD638 pKa = 3.93NPAPVVHH645 pKa = 6.58TATGRR650 pKa = 11.84DD651 pKa = 3.82YY652 pKa = 11.33PDD654 pKa = 4.02ALAGSALAGFQGVPVVLSRR673 pKa = 11.84PDD675 pKa = 3.28GLPNVVRR682 pKa = 11.84DD683 pKa = 3.9GMVALAPEE691 pKa = 4.33SAVILGGEE699 pKa = 4.22GALNDD704 pKa = 3.92DD705 pKa = 3.52VRR707 pKa = 11.84TEE709 pKa = 3.88IEE711 pKa = 4.34ALFEE715 pKa = 5.15DD716 pKa = 5.04SSQTYY721 pKa = 9.54PGISSGMGVGPAPLVNSVDD740 pKa = 3.11IVAADD745 pKa = 3.54STVADD750 pKa = 4.97AEE752 pKa = 4.35LCLPGSLDD760 pKa = 3.66AAGAADD766 pKa = 4.43NIVICTRR773 pKa = 11.84GTNARR778 pKa = 11.84VEE780 pKa = 4.15KK781 pKa = 10.81SEE783 pKa = 4.17TVAEE787 pKa = 4.22AGGIGMVLANNNNEE801 pKa = 3.76EE802 pKa = 4.17SLNGDD807 pKa = 3.78FHH809 pKa = 7.15SVPTIHH815 pKa = 7.27VDD817 pKa = 3.12GTTGDD822 pKa = 3.79AIKK825 pKa = 10.2AYY827 pKa = 8.88EE828 pKa = 4.38ASDD831 pKa = 4.03PNPTATISATGEE843 pKa = 4.38GPEE846 pKa = 4.0LTYY849 pKa = 10.97PEE851 pKa = 4.14MAGFSSYY858 pKa = 11.08GPASAGGGDD867 pKa = 4.33LLKK870 pKa = 10.49PDD872 pKa = 3.24ITAPGVDD879 pKa = 3.98VIAAVSPAGNGGNNFDD895 pKa = 4.52SYY897 pKa = 11.67SGTSMSAPHH906 pKa = 6.56IAGLAALMMQDD917 pKa = 3.41NPEE920 pKa = 4.13WSPMAVKK927 pKa = 10.49SSMMTTADD935 pKa = 3.49PTNSEE940 pKa = 4.34GEE942 pKa = 4.29LINYY946 pKa = 8.95AGAPASPLHH955 pKa = 5.84YY956 pKa = 10.35GSGEE960 pKa = 4.16VEE962 pKa = 3.93PGAAYY967 pKa = 7.64GTPLVYY973 pKa = 10.74DD974 pKa = 3.77SDD976 pKa = 4.51VIDD979 pKa = 4.25WIGYY983 pKa = 8.52ACSIGQWQLIGGAADD998 pKa = 3.71CADD1001 pKa = 4.75LGEE1004 pKa = 5.07SDD1006 pKa = 5.08PSDD1009 pKa = 3.68LNYY1012 pKa = 10.47PSIAIGDD1019 pKa = 3.86LGGTQTITRR1028 pKa = 11.84TVTDD1032 pKa = 3.65ASGEE1036 pKa = 4.3GGTFTAEE1043 pKa = 3.78VEE1045 pKa = 4.24APPGIDD1051 pKa = 2.86AVVEE1055 pKa = 4.03PSTITVEE1062 pKa = 4.25PGGTATFEE1070 pKa = 4.42VTFTTTDD1077 pKa = 3.1AEE1079 pKa = 4.32LGEE1082 pKa = 4.33WTFGSLTWTGPGEE1095 pKa = 4.28DD1096 pKa = 3.71VRR1098 pKa = 11.84SPIAINPVAMGAPAEE1113 pKa = 4.07IRR1115 pKa = 11.84GEE1117 pKa = 4.11GTEE1120 pKa = 4.09GSEE1123 pKa = 4.33TFEE1126 pKa = 4.12VTPGFSGEE1134 pKa = 4.18LTSVVHH1140 pKa = 6.81GLMASDD1146 pKa = 3.74VTDD1149 pKa = 4.82LDD1151 pKa = 3.95VTSDD1155 pKa = 3.62GPAGGDD1161 pKa = 3.75GIADD1165 pKa = 3.36AVVPVTVPEE1174 pKa = 4.24GTSAIRR1180 pKa = 11.84VEE1182 pKa = 4.54VVEE1185 pKa = 5.32SEE1187 pKa = 4.07WTPDD1191 pKa = 3.75GLDD1194 pKa = 3.45LDD1196 pKa = 5.63LYY1198 pKa = 11.12LADD1201 pKa = 3.85SEE1203 pKa = 5.11GNVVGQSAAGGSDD1216 pKa = 3.23EE1217 pKa = 5.34SVTTVAPAPGDD1228 pKa = 3.52YY1229 pKa = 10.66LVAIDD1234 pKa = 4.46FWSGAEE1240 pKa = 4.21GEE1242 pKa = 4.66VASGPLHH1249 pKa = 6.96VFMPSADD1256 pKa = 3.98EE1257 pKa = 4.69GNMTVTPSPVAVTAGVPLDD1276 pKa = 4.18LTAAWTGLDD1285 pKa = 3.01AATRR1289 pKa = 11.84YY1290 pKa = 10.15LGVIGYY1296 pKa = 7.48STGGEE1301 pKa = 4.08EE1302 pKa = 4.29VASTLVSVNTPP1313 pKa = 3.03
MM1 pKa = 7.12HH2 pKa = 6.79QPSARR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84TRR11 pKa = 11.84LLALAGAGAVIAPLALVAPSAWANPDD37 pKa = 3.16LTVNEE42 pKa = 4.7DD43 pKa = 2.74GTYY46 pKa = 10.44IVMLADD52 pKa = 3.7SSLAAYY58 pKa = 10.12DD59 pKa = 4.07GGVQGIPATKK69 pKa = 9.11PAEE72 pKa = 4.25GEE74 pKa = 4.19KK75 pKa = 10.88LSTTTAAAEE84 pKa = 4.59DD85 pKa = 3.73YY86 pKa = 8.93TAYY89 pKa = 11.08LDD91 pKa = 3.79RR92 pKa = 11.84QQNAVLDD99 pKa = 4.21SVGLDD104 pKa = 3.18RR105 pKa = 11.84ADD107 pKa = 3.5AVYY110 pKa = 9.22TYY112 pKa = 9.72STAFNGFTADD122 pKa = 3.48LTGSQVSALRR132 pKa = 11.84KK133 pKa = 8.91DD134 pKa = 3.78PKK136 pKa = 10.71VAAVWEE142 pKa = 4.24NEE144 pKa = 3.44VRR146 pKa = 11.84YY147 pKa = 8.16TDD149 pKa = 4.23TITTPDD155 pKa = 3.76YY156 pKa = 11.38LGLTGDD162 pKa = 3.78DD163 pKa = 4.36GVWNTQFGGDD173 pKa = 3.63ANAGSGIVVGIIDD186 pKa = 3.49TGFWPEE192 pKa = 4.06NPSFAALPGDD202 pKa = 4.35PEE204 pKa = 5.42PPADD208 pKa = 3.49WNGEE212 pKa = 4.25CVEE215 pKa = 4.84GDD217 pKa = 3.74SADD220 pKa = 3.63PADD223 pKa = 4.68NVTCNSKK230 pKa = 10.85VIGARR235 pKa = 11.84FYY237 pKa = 10.81PDD239 pKa = 3.25TNDD242 pKa = 2.93TSFDD246 pKa = 3.75FEE248 pKa = 4.87SPRR251 pKa = 11.84DD252 pKa = 4.0TNGHH256 pKa = 5.84GSHH259 pKa = 6.62VGGTSAGNVDD269 pKa = 3.8VPMEE273 pKa = 4.3INGVEE278 pKa = 4.02LGEE281 pKa = 4.37GSGMAPAAHH290 pKa = 6.54LAFYY294 pKa = 9.93KK295 pKa = 10.76ALWQTEE301 pKa = 4.16DD302 pKa = 3.76GNGSGTSAGLVAAIDD317 pKa = 3.86DD318 pKa = 4.13AVADD322 pKa = 4.22GVDD325 pKa = 3.67VINYY329 pKa = 8.3SVSGSSDD336 pKa = 3.52YY337 pKa = 11.53VVTADD342 pKa = 4.17EE343 pKa = 5.11LAFLDD348 pKa = 4.1AANAGVFVSTSAGNSGDD365 pKa = 3.76TVGEE369 pKa = 3.8SSVAHH374 pKa = 5.73NSPWTMTVAASTHH387 pKa = 6.34DD388 pKa = 3.59RR389 pKa = 11.84TNSAEE394 pKa = 3.99VEE396 pKa = 4.3LGGGAPVTRR405 pKa = 11.84ISGTNRR411 pKa = 11.84YY412 pKa = 7.67DD413 pKa = 3.11TAAQIAAAYY422 pKa = 8.52PEE424 pKa = 4.84GVDD427 pKa = 3.21TVYY430 pKa = 10.64IATGNQFADD439 pKa = 3.78ALSGAAAASQGLVPANGLDD458 pKa = 3.77QPAVAPDD465 pKa = 4.26GSPAPVLLTKK475 pKa = 10.35VDD477 pKa = 4.95EE478 pKa = 4.76IPSATMEE485 pKa = 4.38ALEE488 pKa = 5.81SIDD491 pKa = 3.56PDD493 pKa = 3.88NIVVLGGEE501 pKa = 4.41VAVSDD506 pKa = 4.59DD507 pKa = 4.02VEE509 pKa = 4.7DD510 pKa = 4.14ALGDD514 pKa = 3.73YY515 pKa = 8.54GTVTRR520 pKa = 11.84VAGANRR526 pKa = 11.84YY527 pKa = 6.69EE528 pKa = 4.3TAALVAEE535 pKa = 4.58QYY537 pKa = 11.53GEE539 pKa = 3.93VDD541 pKa = 3.17HH542 pKa = 7.02VYY544 pKa = 10.61VATGQDD550 pKa = 3.19EE551 pKa = 4.73AFPDD555 pKa = 3.94ALSGSALAGSEE566 pKa = 4.19GVPVLLTKK574 pKa = 10.65SDD576 pKa = 3.88MVPGAVSDD584 pKa = 3.96ALEE587 pKa = 4.21AMGDD591 pKa = 3.87PEE593 pKa = 4.19VHH595 pKa = 6.62VIGGSAAVSEE605 pKa = 5.23DD606 pKa = 3.26VFEE609 pKa = 5.02DD610 pKa = 3.31LGGSEE615 pKa = 4.52RR616 pKa = 11.84LSGTDD621 pKa = 2.83RR622 pKa = 11.84YY623 pKa = 8.48GTSVAVAEE631 pKa = 4.25QFGYY635 pKa = 10.81DD636 pKa = 3.81ADD638 pKa = 3.93NPAPVVHH645 pKa = 6.58TATGRR650 pKa = 11.84DD651 pKa = 3.82YY652 pKa = 11.33PDD654 pKa = 4.02ALAGSALAGFQGVPVVLSRR673 pKa = 11.84PDD675 pKa = 3.28GLPNVVRR682 pKa = 11.84DD683 pKa = 3.9GMVALAPEE691 pKa = 4.33SAVILGGEE699 pKa = 4.22GALNDD704 pKa = 3.92DD705 pKa = 3.52VRR707 pKa = 11.84TEE709 pKa = 3.88IEE711 pKa = 4.34ALFEE715 pKa = 5.15DD716 pKa = 5.04SSQTYY721 pKa = 9.54PGISSGMGVGPAPLVNSVDD740 pKa = 3.11IVAADD745 pKa = 3.54STVADD750 pKa = 4.97AEE752 pKa = 4.35LCLPGSLDD760 pKa = 3.66AAGAADD766 pKa = 4.43NIVICTRR773 pKa = 11.84GTNARR778 pKa = 11.84VEE780 pKa = 4.15KK781 pKa = 10.81SEE783 pKa = 4.17TVAEE787 pKa = 4.22AGGIGMVLANNNNEE801 pKa = 3.76EE802 pKa = 4.17SLNGDD807 pKa = 3.78FHH809 pKa = 7.15SVPTIHH815 pKa = 7.27VDD817 pKa = 3.12GTTGDD822 pKa = 3.79AIKK825 pKa = 10.2AYY827 pKa = 8.88EE828 pKa = 4.38ASDD831 pKa = 4.03PNPTATISATGEE843 pKa = 4.38GPEE846 pKa = 4.0LTYY849 pKa = 10.97PEE851 pKa = 4.14MAGFSSYY858 pKa = 11.08GPASAGGGDD867 pKa = 4.33LLKK870 pKa = 10.49PDD872 pKa = 3.24ITAPGVDD879 pKa = 3.98VIAAVSPAGNGGNNFDD895 pKa = 4.52SYY897 pKa = 11.67SGTSMSAPHH906 pKa = 6.56IAGLAALMMQDD917 pKa = 3.41NPEE920 pKa = 4.13WSPMAVKK927 pKa = 10.49SSMMTTADD935 pKa = 3.49PTNSEE940 pKa = 4.34GEE942 pKa = 4.29LINYY946 pKa = 8.95AGAPASPLHH955 pKa = 5.84YY956 pKa = 10.35GSGEE960 pKa = 4.16VEE962 pKa = 3.93PGAAYY967 pKa = 7.64GTPLVYY973 pKa = 10.74DD974 pKa = 3.77SDD976 pKa = 4.51VIDD979 pKa = 4.25WIGYY983 pKa = 8.52ACSIGQWQLIGGAADD998 pKa = 3.71CADD1001 pKa = 4.75LGEE1004 pKa = 5.07SDD1006 pKa = 5.08PSDD1009 pKa = 3.68LNYY1012 pKa = 10.47PSIAIGDD1019 pKa = 3.86LGGTQTITRR1028 pKa = 11.84TVTDD1032 pKa = 3.65ASGEE1036 pKa = 4.3GGTFTAEE1043 pKa = 3.78VEE1045 pKa = 4.24APPGIDD1051 pKa = 2.86AVVEE1055 pKa = 4.03PSTITVEE1062 pKa = 4.25PGGTATFEE1070 pKa = 4.42VTFTTTDD1077 pKa = 3.1AEE1079 pKa = 4.32LGEE1082 pKa = 4.33WTFGSLTWTGPGEE1095 pKa = 4.28DD1096 pKa = 3.71VRR1098 pKa = 11.84SPIAINPVAMGAPAEE1113 pKa = 4.07IRR1115 pKa = 11.84GEE1117 pKa = 4.11GTEE1120 pKa = 4.09GSEE1123 pKa = 4.33TFEE1126 pKa = 4.12VTPGFSGEE1134 pKa = 4.18LTSVVHH1140 pKa = 6.81GLMASDD1146 pKa = 3.74VTDD1149 pKa = 4.82LDD1151 pKa = 3.95VTSDD1155 pKa = 3.62GPAGGDD1161 pKa = 3.75GIADD1165 pKa = 3.36AVVPVTVPEE1174 pKa = 4.24GTSAIRR1180 pKa = 11.84VEE1182 pKa = 4.54VVEE1185 pKa = 5.32SEE1187 pKa = 4.07WTPDD1191 pKa = 3.75GLDD1194 pKa = 3.45LDD1196 pKa = 5.63LYY1198 pKa = 11.12LADD1201 pKa = 3.85SEE1203 pKa = 5.11GNVVGQSAAGGSDD1216 pKa = 3.23EE1217 pKa = 5.34SVTTVAPAPGDD1228 pKa = 3.52YY1229 pKa = 10.66LVAIDD1234 pKa = 4.46FWSGAEE1240 pKa = 4.21GEE1242 pKa = 4.66VASGPLHH1249 pKa = 6.96VFMPSADD1256 pKa = 3.98EE1257 pKa = 4.69GNMTVTPSPVAVTAGVPLDD1276 pKa = 4.18LTAAWTGLDD1285 pKa = 3.01AATRR1289 pKa = 11.84YY1290 pKa = 10.15LGVIGYY1296 pKa = 7.48STGGEE1301 pKa = 4.08EE1302 pKa = 4.29VASTLVSVNTPP1313 pKa = 3.03
Molecular weight: 132.99 kDa
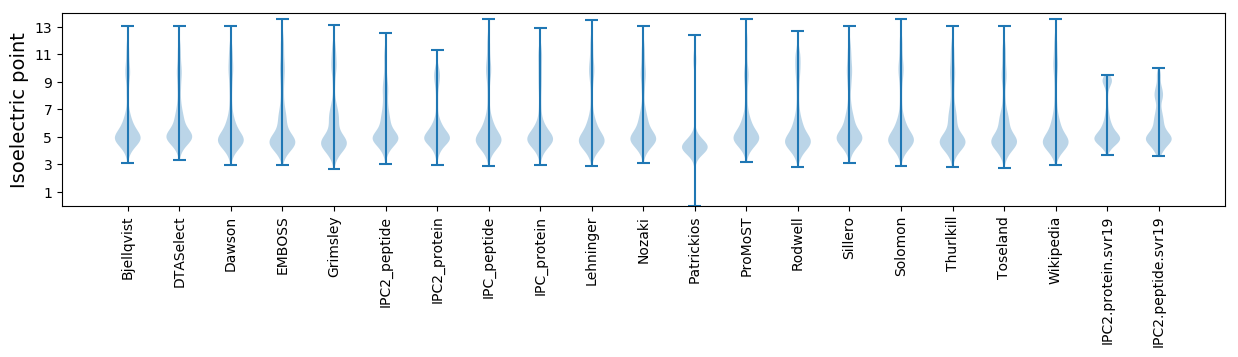
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B1NCW4|A0A1B1NCW4_9MICO Foldase YidC OS=Serinicoccus hydrothermalis OX=1758689 GN=SGUI_1876 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84GILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SALSAA45 pKa = 3.8
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84GILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SALSAA45 pKa = 3.8
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1072852 |
37 |
2118 |
325.1 |
34.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.837 ± 0.058 | 0.645 ± 0.011 |
6.477 ± 0.042 | 6.351 ± 0.045 |
2.527 ± 0.026 | 9.671 ± 0.04 |
2.236 ± 0.024 | 3.068 ± 0.032 |
1.419 ± 0.029 | 10.583 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.907 ± 0.019 | 1.427 ± 0.025 |
5.728 ± 0.035 | 3.11 ± 0.024 |
7.847 ± 0.053 | 5.11 ± 0.031 |
6.058 ± 0.033 | 9.548 ± 0.04 |
1.606 ± 0.022 | 1.843 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
