
Dentipellis sp. KUC8613
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Russulales; Hericiaceae; Dentipellis; unclassified Dentipellis
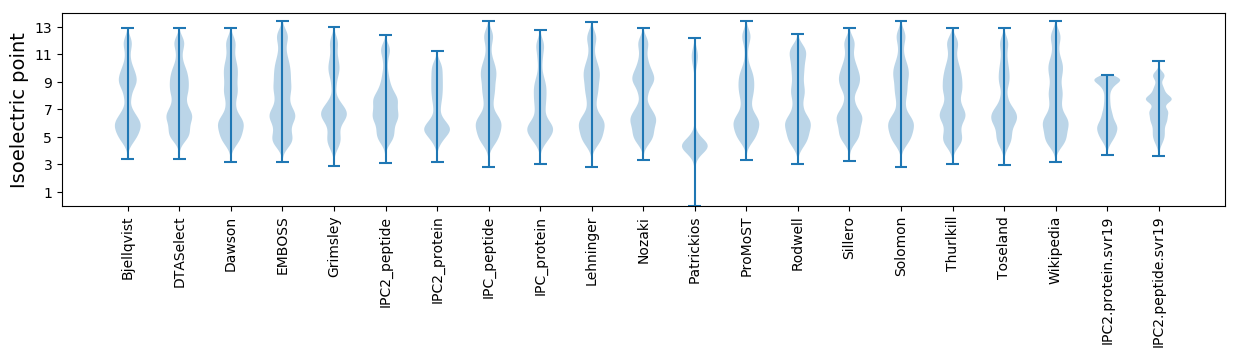
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
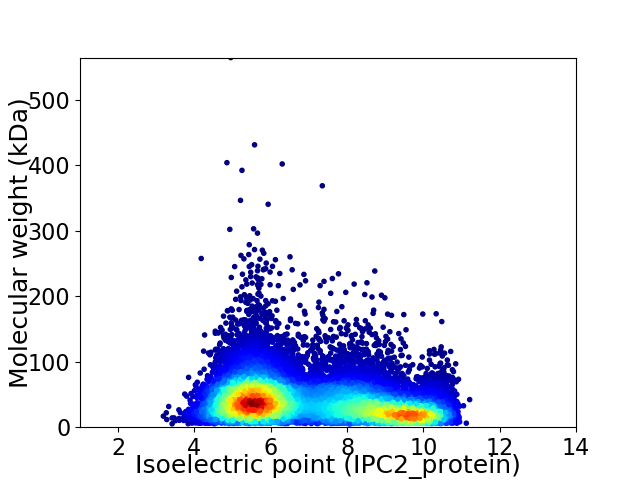
Virtual 2D-PAGE plot for 14287 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B1QFU5|A0A5B1QFU5_9AGAM Uncharacterized protein OS=Dentipellis sp. KUC8613 OX=1883078 GN=DENSPDRAFT_833366 PE=4 SV=1
MM1 pKa = 7.34FATLFTVTLLSAGAIKK17 pKa = 10.46GALAGLAINSPTLVQCEE34 pKa = 4.09DD35 pKa = 3.6AHH37 pKa = 6.88VSWASTKK44 pKa = 10.43GPYY47 pKa = 10.08NLIVTPGDD55 pKa = 3.95EE56 pKa = 4.39PCGDD60 pKa = 4.68AIVDD64 pKa = 4.35LGDD67 pKa = 3.93HH68 pKa = 7.39DD69 pKa = 4.64GTTMTYY75 pKa = 10.74NVALPAGQKK84 pKa = 10.12VLLSLQDD91 pKa = 3.73ANGDD95 pKa = 3.93EE96 pKa = 4.53AWSQEE101 pKa = 3.93LTVQPGSNSACLPASLQPSSSIATLSAASSSVKK134 pKa = 9.85FASSSAPFSLSSTVVPLTAVTPATTISALALPSDD168 pKa = 4.25SAAAAPSDD176 pKa = 4.17TIGGDD181 pKa = 3.57DD182 pKa = 3.63SDD184 pKa = 4.41APSVVGGAANAASNPFSGAPAMHH207 pKa = 6.31QLYY210 pKa = 8.28TPAMFLSALGAAFFVALL227 pKa = 4.35
MM1 pKa = 7.34FATLFTVTLLSAGAIKK17 pKa = 10.46GALAGLAINSPTLVQCEE34 pKa = 4.09DD35 pKa = 3.6AHH37 pKa = 6.88VSWASTKK44 pKa = 10.43GPYY47 pKa = 10.08NLIVTPGDD55 pKa = 3.95EE56 pKa = 4.39PCGDD60 pKa = 4.68AIVDD64 pKa = 4.35LGDD67 pKa = 3.93HH68 pKa = 7.39DD69 pKa = 4.64GTTMTYY75 pKa = 10.74NVALPAGQKK84 pKa = 10.12VLLSLQDD91 pKa = 3.73ANGDD95 pKa = 3.93EE96 pKa = 4.53AWSQEE101 pKa = 3.93LTVQPGSNSACLPASLQPSSSIATLSAASSSVKK134 pKa = 9.85FASSSAPFSLSSTVVPLTAVTPATTISALALPSDD168 pKa = 4.25SAAAAPSDD176 pKa = 4.17TIGGDD181 pKa = 3.57DD182 pKa = 3.63SDD184 pKa = 4.41APSVVGGAANAASNPFSGAPAMHH207 pKa = 6.31QLYY210 pKa = 8.28TPAMFLSALGAAFFVALL227 pKa = 4.35
Molecular weight: 22.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B1RER3|A0A5B1RER3_9AGAM Uncharacterized protein OS=Dentipellis sp. KUC8613 OX=1883078 GN=DENSPDRAFT_887317 PE=4 SV=1
MM1 pKa = 6.33QTRR4 pKa = 11.84LRR6 pKa = 11.84LVLSLSRR13 pKa = 11.84CLVPSLPPSRR23 pKa = 11.84PLRR26 pKa = 11.84AVAPPHH32 pKa = 5.43TLVVSSHH39 pKa = 5.32HH40 pKa = 6.18RR41 pKa = 11.84AVILSRR47 pKa = 11.84HH48 pKa = 5.44RR49 pKa = 11.84ASLAHH54 pKa = 6.85LPRR57 pKa = 11.84PASTLVTSSSRR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84ILVCRR75 pKa = 11.84HH76 pKa = 5.03RR77 pKa = 11.84TLVAPSFPRR86 pKa = 11.84AVALSRR92 pKa = 11.84SSRR95 pKa = 11.84APPAVSMLSPRR106 pKa = 11.84RR107 pKa = 11.84VVVASSRR114 pKa = 11.84PAPPALLVPLPPLQPPPCRR133 pKa = 11.84APHH136 pKa = 6.17PRR138 pKa = 11.84VAPSPSVASSRR149 pKa = 11.84PCRR152 pKa = 11.84ARR154 pKa = 11.84RR155 pKa = 11.84VRR157 pKa = 11.84VAPVASVSRR166 pKa = 11.84PSRR169 pKa = 11.84PSRR172 pKa = 11.84PSRR175 pKa = 11.84PRR177 pKa = 11.84LAVAPILRR185 pKa = 11.84PSRR188 pKa = 11.84RR189 pKa = 11.84SRR191 pKa = 11.84HH192 pKa = 4.6RR193 pKa = 11.84ARR195 pKa = 11.84PRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84AVAPVVPLCRR209 pKa = 11.84RR210 pKa = 11.84ARR212 pKa = 11.84RR213 pKa = 11.84AVAALPLSALAPPSRR228 pKa = 11.84PSHH231 pKa = 5.54AHH233 pKa = 5.6HH234 pKa = 6.81VPSAHH239 pKa = 6.28AQPSRR244 pKa = 11.84PSRR247 pKa = 11.84VRR249 pKa = 11.84RR250 pKa = 11.84ARR252 pKa = 11.84HH253 pKa = 5.49APRR256 pKa = 11.84SVPRR260 pKa = 11.84HH261 pKa = 4.69ARR263 pKa = 11.84RR264 pKa = 11.84ARR266 pKa = 11.84RR267 pKa = 11.84TPFASRR273 pKa = 11.84PSRR276 pKa = 11.84HH277 pKa = 5.38RR278 pKa = 11.84ASRR281 pKa = 11.84RR282 pKa = 11.84LRR284 pKa = 11.84AVVPSRR290 pKa = 11.84PSRR293 pKa = 11.84VRR295 pKa = 11.84RR296 pKa = 11.84AVATFAPLSAIAPPSRR312 pKa = 11.84PAHH315 pKa = 5.73AHH317 pKa = 5.72HH318 pKa = 7.09APGAIAPVAAFAPSRR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84AVAPVARR342 pKa = 11.84LSRR345 pKa = 11.84PIRR348 pKa = 11.84AAGVMPSRR356 pKa = 11.84RR357 pKa = 11.84RR358 pKa = 11.84HH359 pKa = 4.87ALARR363 pKa = 11.84PARR366 pKa = 11.84PRR368 pKa = 11.84AHH370 pKa = 6.58FVPVLHH376 pKa = 6.7LHH378 pKa = 5.68MDD380 pKa = 3.97VYY382 pKa = 11.68VLLL385 pKa = 4.72
MM1 pKa = 6.33QTRR4 pKa = 11.84LRR6 pKa = 11.84LVLSLSRR13 pKa = 11.84CLVPSLPPSRR23 pKa = 11.84PLRR26 pKa = 11.84AVAPPHH32 pKa = 5.43TLVVSSHH39 pKa = 5.32HH40 pKa = 6.18RR41 pKa = 11.84AVILSRR47 pKa = 11.84HH48 pKa = 5.44RR49 pKa = 11.84ASLAHH54 pKa = 6.85LPRR57 pKa = 11.84PASTLVTSSSRR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84ILVCRR75 pKa = 11.84HH76 pKa = 5.03RR77 pKa = 11.84TLVAPSFPRR86 pKa = 11.84AVALSRR92 pKa = 11.84SSRR95 pKa = 11.84APPAVSMLSPRR106 pKa = 11.84RR107 pKa = 11.84VVVASSRR114 pKa = 11.84PAPPALLVPLPPLQPPPCRR133 pKa = 11.84APHH136 pKa = 6.17PRR138 pKa = 11.84VAPSPSVASSRR149 pKa = 11.84PCRR152 pKa = 11.84ARR154 pKa = 11.84RR155 pKa = 11.84VRR157 pKa = 11.84VAPVASVSRR166 pKa = 11.84PSRR169 pKa = 11.84PSRR172 pKa = 11.84PSRR175 pKa = 11.84PRR177 pKa = 11.84LAVAPILRR185 pKa = 11.84PSRR188 pKa = 11.84RR189 pKa = 11.84SRR191 pKa = 11.84HH192 pKa = 4.6RR193 pKa = 11.84ARR195 pKa = 11.84PRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84AVAPVVPLCRR209 pKa = 11.84RR210 pKa = 11.84ARR212 pKa = 11.84RR213 pKa = 11.84AVAALPLSALAPPSRR228 pKa = 11.84PSHH231 pKa = 5.54AHH233 pKa = 5.6HH234 pKa = 6.81VPSAHH239 pKa = 6.28AQPSRR244 pKa = 11.84PSRR247 pKa = 11.84VRR249 pKa = 11.84RR250 pKa = 11.84ARR252 pKa = 11.84HH253 pKa = 5.49APRR256 pKa = 11.84SVPRR260 pKa = 11.84HH261 pKa = 4.69ARR263 pKa = 11.84RR264 pKa = 11.84ARR266 pKa = 11.84RR267 pKa = 11.84TPFASRR273 pKa = 11.84PSRR276 pKa = 11.84HH277 pKa = 5.38RR278 pKa = 11.84ASRR281 pKa = 11.84RR282 pKa = 11.84LRR284 pKa = 11.84AVVPSRR290 pKa = 11.84PSRR293 pKa = 11.84VRR295 pKa = 11.84RR296 pKa = 11.84AVATFAPLSAIAPPSRR312 pKa = 11.84PAHH315 pKa = 5.73AHH317 pKa = 5.72HH318 pKa = 7.09APGAIAPVAAFAPSRR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84AVAPVARR342 pKa = 11.84LSRR345 pKa = 11.84PIRR348 pKa = 11.84AAGVMPSRR356 pKa = 11.84RR357 pKa = 11.84RR358 pKa = 11.84HH359 pKa = 4.87ALARR363 pKa = 11.84PARR366 pKa = 11.84PRR368 pKa = 11.84AHH370 pKa = 6.58FVPVLHH376 pKa = 6.7LHH378 pKa = 5.68MDD380 pKa = 3.97VYY382 pKa = 11.68VLLL385 pKa = 4.72
Molecular weight: 42.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5785676 |
49 |
5066 |
405.0 |
44.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.826 ± 0.026 | 1.432 ± 0.012 |
5.317 ± 0.019 | 5.521 ± 0.025 |
3.516 ± 0.014 | 6.459 ± 0.023 |
2.752 ± 0.011 | 4.3 ± 0.016 |
4.016 ± 0.02 | 9.138 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.004 ± 0.009 | 2.977 ± 0.013 |
7.494 ± 0.037 | 3.558 ± 0.015 |
7.051 ± 0.031 | 8.549 ± 0.028 |
5.866 ± 0.014 | 6.346 ± 0.016 |
1.405 ± 0.008 | 2.473 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
