
Methanolobus tindarius DSM 2278
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; Methanosarcinaceae; Methanolobus; Methanolobus tindarius
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
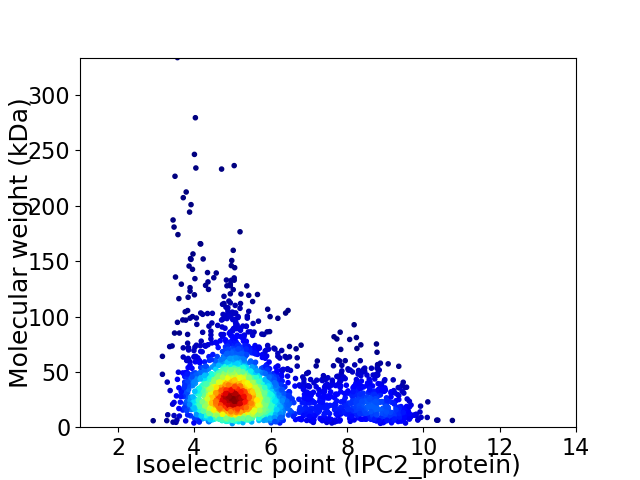
Virtual 2D-PAGE plot for 2912 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W9DN22|W9DN22_METTI Uncharacterized protein OS=Methanolobus tindarius DSM 2278 OX=1090322 GN=MettiDRAFT_0754 PE=4 SV=1
MM1 pKa = 7.16VKK3 pKa = 10.1EE4 pKa = 4.37KK5 pKa = 10.55VHH7 pKa = 7.86IYY9 pKa = 10.32IIFPTFLCLVMSLSTPAYY27 pKa = 9.29AAGDD31 pKa = 3.4ITAVRR36 pKa = 11.84EE37 pKa = 3.91ISNMNVTPGEE47 pKa = 4.23TFNVSVTLKK56 pKa = 10.27MNADD60 pKa = 3.81VIGPLLRR67 pKa = 11.84EE68 pKa = 4.07TLPDD72 pKa = 2.77GWTIGSLDD80 pKa = 3.54NDD82 pKa = 3.21GSLFNVNTTEE92 pKa = 4.21WAWVSMLQSGEE103 pKa = 4.33SKK105 pKa = 8.74TVEE108 pKa = 4.05YY109 pKa = 11.03SVMVPDD115 pKa = 4.38DD116 pKa = 3.94TDD118 pKa = 4.84DD119 pKa = 3.62GVYY122 pKa = 10.12TISGYY127 pKa = 8.99VAPEE131 pKa = 4.0AYY133 pKa = 10.41NYY135 pKa = 8.57ITVLGDD141 pKa = 3.38TQVTVSSEE149 pKa = 3.88TSTPEE154 pKa = 3.7VIADD158 pKa = 4.01FTANITSGKK167 pKa = 10.02VPLTVKK173 pKa = 9.65FTDD176 pKa = 3.1KK177 pKa = 10.34STNATSWQWDD187 pKa = 3.48FDD189 pKa = 4.13GDD191 pKa = 3.66NSIDD195 pKa = 3.67STEE198 pKa = 3.92QNPVYY203 pKa = 9.66TYY205 pKa = 10.86QSAGTYY211 pKa = 6.62TVKK214 pKa = 10.87LISSSNSDD222 pKa = 2.91SDD224 pKa = 4.71VEE226 pKa = 4.48TKK228 pKa = 10.08TDD230 pKa = 3.88YY231 pKa = 10.09ITIKK235 pKa = 10.64SGGGSSGTSLKK246 pKa = 10.83ADD248 pKa = 3.39FTVNITSGQAPLSVQFTDD266 pKa = 3.42QSSGNVSAWQWDD278 pKa = 3.63FDD280 pKa = 4.16GDD282 pKa = 3.96NVIDD286 pKa = 4.0STEE289 pKa = 3.75QDD291 pKa = 3.3PVHH294 pKa = 6.79TYY296 pKa = 9.56TEE298 pKa = 4.2NGSYY302 pKa = 7.97TVKK305 pKa = 10.88LIVYY309 pKa = 9.79DD310 pKa = 4.39DD311 pKa = 4.91DD312 pKa = 5.17SNSDD316 pKa = 3.31ALTEE320 pKa = 3.91VAYY323 pKa = 8.97ITVSDD328 pKa = 4.33EE329 pKa = 4.46EE330 pKa = 6.21DD331 pKa = 3.66DD332 pKa = 4.89DD333 pKa = 5.27SYY335 pKa = 12.22NSYY338 pKa = 10.67EE339 pKa = 4.01ATSVSLSATIIPSISIEE356 pKa = 4.16VTPGTMNFGTLSAGEE371 pKa = 4.18VSDD374 pKa = 4.04EE375 pKa = 4.13HH376 pKa = 6.24TLHH379 pKa = 6.67IKK381 pKa = 10.63NKK383 pKa = 8.48GATDD387 pKa = 3.57ATVTAQVTDD396 pKa = 3.66VAKK399 pKa = 10.64DD400 pKa = 3.61LYY402 pKa = 11.52VDD404 pKa = 4.05GLMLNSGTWDD414 pKa = 3.66GYY416 pKa = 9.51SNVVEE421 pKa = 4.47SEE423 pKa = 4.09TTEE426 pKa = 4.15TVQASLNVPDD436 pKa = 4.95DD437 pKa = 4.04FIGIGSMEE445 pKa = 4.22GKK447 pKa = 10.01LVFWAEE453 pKa = 3.78ISS455 pKa = 3.43
MM1 pKa = 7.16VKK3 pKa = 10.1EE4 pKa = 4.37KK5 pKa = 10.55VHH7 pKa = 7.86IYY9 pKa = 10.32IIFPTFLCLVMSLSTPAYY27 pKa = 9.29AAGDD31 pKa = 3.4ITAVRR36 pKa = 11.84EE37 pKa = 3.91ISNMNVTPGEE47 pKa = 4.23TFNVSVTLKK56 pKa = 10.27MNADD60 pKa = 3.81VIGPLLRR67 pKa = 11.84EE68 pKa = 4.07TLPDD72 pKa = 2.77GWTIGSLDD80 pKa = 3.54NDD82 pKa = 3.21GSLFNVNTTEE92 pKa = 4.21WAWVSMLQSGEE103 pKa = 4.33SKK105 pKa = 8.74TVEE108 pKa = 4.05YY109 pKa = 11.03SVMVPDD115 pKa = 4.38DD116 pKa = 3.94TDD118 pKa = 4.84DD119 pKa = 3.62GVYY122 pKa = 10.12TISGYY127 pKa = 8.99VAPEE131 pKa = 4.0AYY133 pKa = 10.41NYY135 pKa = 8.57ITVLGDD141 pKa = 3.38TQVTVSSEE149 pKa = 3.88TSTPEE154 pKa = 3.7VIADD158 pKa = 4.01FTANITSGKK167 pKa = 10.02VPLTVKK173 pKa = 9.65FTDD176 pKa = 3.1KK177 pKa = 10.34STNATSWQWDD187 pKa = 3.48FDD189 pKa = 4.13GDD191 pKa = 3.66NSIDD195 pKa = 3.67STEE198 pKa = 3.92QNPVYY203 pKa = 9.66TYY205 pKa = 10.86QSAGTYY211 pKa = 6.62TVKK214 pKa = 10.87LISSSNSDD222 pKa = 2.91SDD224 pKa = 4.71VEE226 pKa = 4.48TKK228 pKa = 10.08TDD230 pKa = 3.88YY231 pKa = 10.09ITIKK235 pKa = 10.64SGGGSSGTSLKK246 pKa = 10.83ADD248 pKa = 3.39FTVNITSGQAPLSVQFTDD266 pKa = 3.42QSSGNVSAWQWDD278 pKa = 3.63FDD280 pKa = 4.16GDD282 pKa = 3.96NVIDD286 pKa = 4.0STEE289 pKa = 3.75QDD291 pKa = 3.3PVHH294 pKa = 6.79TYY296 pKa = 9.56TEE298 pKa = 4.2NGSYY302 pKa = 7.97TVKK305 pKa = 10.88LIVYY309 pKa = 9.79DD310 pKa = 4.39DD311 pKa = 4.91DD312 pKa = 5.17SNSDD316 pKa = 3.31ALTEE320 pKa = 3.91VAYY323 pKa = 8.97ITVSDD328 pKa = 4.33EE329 pKa = 4.46EE330 pKa = 6.21DD331 pKa = 3.66DD332 pKa = 4.89DD333 pKa = 5.27SYY335 pKa = 12.22NSYY338 pKa = 10.67EE339 pKa = 4.01ATSVSLSATIIPSISIEE356 pKa = 4.16VTPGTMNFGTLSAGEE371 pKa = 4.18VSDD374 pKa = 4.04EE375 pKa = 4.13HH376 pKa = 6.24TLHH379 pKa = 6.67IKK381 pKa = 10.63NKK383 pKa = 8.48GATDD387 pKa = 3.57ATVTAQVTDD396 pKa = 3.66VAKK399 pKa = 10.64DD400 pKa = 3.61LYY402 pKa = 11.52VDD404 pKa = 4.05GLMLNSGTWDD414 pKa = 3.66GYY416 pKa = 9.51SNVVEE421 pKa = 4.47SEE423 pKa = 4.09TTEE426 pKa = 4.15TVQASLNVPDD436 pKa = 4.95DD437 pKa = 4.04FIGIGSMEE445 pKa = 4.22GKK447 pKa = 10.01LVFWAEE453 pKa = 3.78ISS455 pKa = 3.43
Molecular weight: 49.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W9DP00|W9DP00_METTI Uncharacterized protein OS=Methanolobus tindarius DSM 2278 OX=1090322 GN=MettiDRAFT_0155 PE=4 SV=1
MM1 pKa = 7.14SHH3 pKa = 5.42NTKK6 pKa = 9.9GQKK9 pKa = 8.31TRR11 pKa = 11.84LAKK14 pKa = 10.28AHH16 pKa = 5.91RR17 pKa = 11.84QNHH20 pKa = 5.7RR21 pKa = 11.84VPTWVIIKK29 pKa = 8.85TGRR32 pKa = 11.84KK33 pKa = 8.65VVSHH37 pKa = 6.6PKK39 pKa = 8.12RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.72WRR44 pKa = 11.84RR45 pKa = 11.84SSLDD49 pKa = 3.23VKK51 pKa = 11.09
MM1 pKa = 7.14SHH3 pKa = 5.42NTKK6 pKa = 9.9GQKK9 pKa = 8.31TRR11 pKa = 11.84LAKK14 pKa = 10.28AHH16 pKa = 5.91RR17 pKa = 11.84QNHH20 pKa = 5.7RR21 pKa = 11.84VPTWVIIKK29 pKa = 8.85TGRR32 pKa = 11.84KK33 pKa = 8.65VVSHH37 pKa = 6.6PKK39 pKa = 8.12RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.72WRR44 pKa = 11.84RR45 pKa = 11.84SSLDD49 pKa = 3.23VKK51 pKa = 11.09
Molecular weight: 6.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
910742 |
29 |
3072 |
312.8 |
34.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.761 ± 0.046 | 1.176 ± 0.021 |
6.396 ± 0.042 | 7.686 ± 0.056 |
4.079 ± 0.029 | 6.871 ± 0.044 |
1.786 ± 0.02 | 8.265 ± 0.043 |
6.262 ± 0.055 | 8.808 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.99 ± 0.025 | 4.709 ± 0.048 |
3.565 ± 0.028 | 2.514 ± 0.024 |
3.854 ± 0.035 | 7.125 ± 0.047 |
5.529 ± 0.045 | 7.184 ± 0.041 |
0.879 ± 0.016 | 3.56 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
