
Thielaviopsis punctulata
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Microascales; Ceratocystidaceae; Thielaviopsis
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
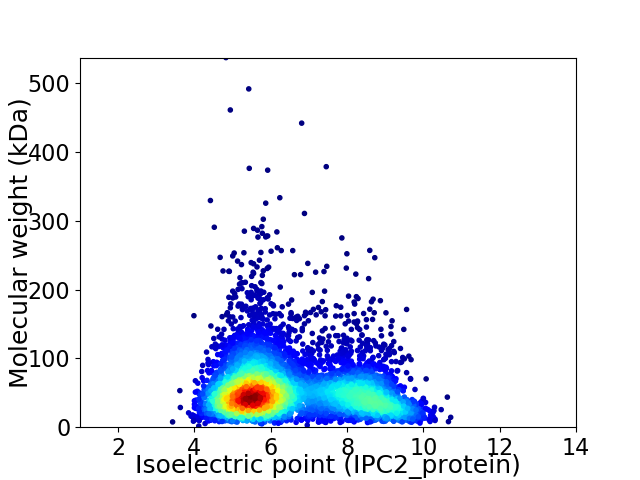
Virtual 2D-PAGE plot for 5254 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F4ZKS4|A0A0F4ZKS4_9PEZI EXS domain-containing protein OS=Thielaviopsis punctulata OX=72032 GN=TD95_005055 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 10.3FPSTAALFLALSGFALAQDD21 pKa = 4.05VSSTPSTSCVSGAACATAQTTMSSVVSSVTASTTTTAAAATSTHH65 pKa = 7.37DD66 pKa = 4.75DD67 pKa = 4.09DD68 pKa = 6.82HH69 pKa = 9.17DD70 pKa = 5.21DD71 pKa = 5.11DD72 pKa = 6.73HH73 pKa = 9.36DD74 pKa = 5.32DD75 pKa = 5.08DD76 pKa = 6.79HH77 pKa = 9.15DD78 pKa = 5.23DD79 pKa = 4.1DD80 pKa = 4.77HH81 pKa = 7.66AAGNGSLAPSPTEE94 pKa = 3.94SVGCEE99 pKa = 3.43PHH101 pKa = 6.71GDD103 pKa = 3.31HH104 pKa = 6.47WHH106 pKa = 6.86CEE108 pKa = 4.09APATAAAATGTASGSATSAGSVQEE132 pKa = 4.48TEE134 pKa = 4.02TATAGGDD141 pKa = 3.54KK142 pKa = 10.91VSVFAGALAIVILAII157 pKa = 4.28
MM1 pKa = 7.68KK2 pKa = 10.3FPSTAALFLALSGFALAQDD21 pKa = 4.05VSSTPSTSCVSGAACATAQTTMSSVVSSVTASTTTTAAAATSTHH65 pKa = 7.37DD66 pKa = 4.75DD67 pKa = 4.09DD68 pKa = 6.82HH69 pKa = 9.17DD70 pKa = 5.21DD71 pKa = 5.11DD72 pKa = 6.73HH73 pKa = 9.36DD74 pKa = 5.32DD75 pKa = 5.08DD76 pKa = 6.79HH77 pKa = 9.15DD78 pKa = 5.23DD79 pKa = 4.1DD80 pKa = 4.77HH81 pKa = 7.66AAGNGSLAPSPTEE94 pKa = 3.94SVGCEE99 pKa = 3.43PHH101 pKa = 6.71GDD103 pKa = 3.31HH104 pKa = 6.47WHH106 pKa = 6.86CEE108 pKa = 4.09APATAAAATGTASGSATSAGSVQEE132 pKa = 4.48TEE134 pKa = 4.02TATAGGDD141 pKa = 3.54KK142 pKa = 10.91VSVFAGALAIVILAII157 pKa = 4.28
Molecular weight: 15.31 kDa
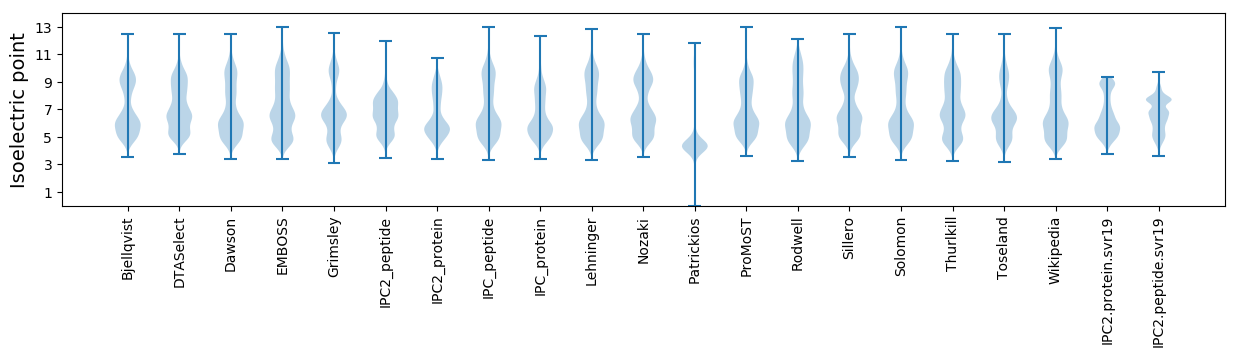
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F4ZFK2|A0A0F4ZFK2_9PEZI MI domain-containing protein OS=Thielaviopsis punctulata OX=72032 GN=TD95_002508 PE=3 SV=1
MM1 pKa = 7.16AQVLVSPSAADD12 pKa = 3.27SAANVAARR20 pKa = 11.84ALLNGASPRR29 pKa = 11.84PAPPPALSAPVPSIPRR45 pKa = 11.84RR46 pKa = 11.84PMTDD50 pKa = 3.2EE51 pKa = 3.52EE52 pKa = 4.17HH53 pKa = 6.41AALVKK58 pKa = 10.43AQRR61 pKa = 11.84AAKK64 pKa = 10.27AGVRR68 pKa = 11.84PPRR71 pKa = 11.84RR72 pKa = 11.84SHH74 pKa = 5.91SASEE78 pKa = 3.98FDD80 pKa = 5.71IEE82 pKa = 4.32QQMPNHH88 pKa = 6.87PPQPRR93 pKa = 11.84YY94 pKa = 10.38LYY96 pKa = 9.95TPVLAPTQASPISPLTPLTPGLPAVMEE123 pKa = 4.34LASPSTPTTPSTNTSLPPQPPMQVRR148 pKa = 11.84TARR151 pKa = 11.84PKK153 pKa = 9.39PKK155 pKa = 9.99KK156 pKa = 8.44PPRR159 pKa = 11.84RR160 pKa = 11.84AFSLMEE166 pKa = 3.99YY167 pKa = 9.79EE168 pKa = 4.6PVVHH172 pKa = 5.76THH174 pKa = 6.23RR175 pKa = 11.84PSNRR179 pKa = 11.84VSLLDD184 pKa = 4.19LPTDD188 pKa = 2.9IHH190 pKa = 6.54FAIFDD195 pKa = 3.88FLDD198 pKa = 5.16PIDD201 pKa = 4.22SVCFGLSSRR210 pKa = 11.84VFYY213 pKa = 9.93PIHH216 pKa = 6.57RR217 pKa = 11.84RR218 pKa = 11.84LHH220 pKa = 5.64GRR222 pKa = 11.84VSLASRR228 pKa = 11.84RR229 pKa = 11.84NGPNTLDD236 pKa = 3.87GPNPARR242 pKa = 11.84PPPRR246 pKa = 11.84SPGLVDD252 pKa = 4.15RR253 pKa = 11.84EE254 pKa = 4.41AIRR257 pKa = 11.84LDD259 pKa = 3.26GQVYY263 pKa = 9.81CRR265 pKa = 11.84KK266 pKa = 9.95CGTSRR271 pKa = 11.84CEE273 pKa = 3.79LHH275 pKa = 5.71SHH277 pKa = 5.35LRR279 pKa = 11.84EE280 pKa = 3.76WMGPDD285 pKa = 3.39RR286 pKa = 11.84EE287 pKa = 4.38YY288 pKa = 11.33CSVSRR293 pKa = 11.84KK294 pKa = 9.24FGKK297 pKa = 9.81KK298 pKa = 9.24APAGARR304 pKa = 11.84EE305 pKa = 4.19SCYY308 pKa = 10.35KK309 pKa = 10.5RR310 pKa = 11.84SPKK313 pKa = 10.29NPSACGRR320 pKa = 11.84HH321 pKa = 4.71TPRR324 pKa = 11.84MRR326 pKa = 11.84ASAPVSPKK334 pKa = 10.35LQAVQMAA341 pKa = 4.15
MM1 pKa = 7.16AQVLVSPSAADD12 pKa = 3.27SAANVAARR20 pKa = 11.84ALLNGASPRR29 pKa = 11.84PAPPPALSAPVPSIPRR45 pKa = 11.84RR46 pKa = 11.84PMTDD50 pKa = 3.2EE51 pKa = 3.52EE52 pKa = 4.17HH53 pKa = 6.41AALVKK58 pKa = 10.43AQRR61 pKa = 11.84AAKK64 pKa = 10.27AGVRR68 pKa = 11.84PPRR71 pKa = 11.84RR72 pKa = 11.84SHH74 pKa = 5.91SASEE78 pKa = 3.98FDD80 pKa = 5.71IEE82 pKa = 4.32QQMPNHH88 pKa = 6.87PPQPRR93 pKa = 11.84YY94 pKa = 10.38LYY96 pKa = 9.95TPVLAPTQASPISPLTPLTPGLPAVMEE123 pKa = 4.34LASPSTPTTPSTNTSLPPQPPMQVRR148 pKa = 11.84TARR151 pKa = 11.84PKK153 pKa = 9.39PKK155 pKa = 9.99KK156 pKa = 8.44PPRR159 pKa = 11.84RR160 pKa = 11.84AFSLMEE166 pKa = 3.99YY167 pKa = 9.79EE168 pKa = 4.6PVVHH172 pKa = 5.76THH174 pKa = 6.23RR175 pKa = 11.84PSNRR179 pKa = 11.84VSLLDD184 pKa = 4.19LPTDD188 pKa = 2.9IHH190 pKa = 6.54FAIFDD195 pKa = 3.88FLDD198 pKa = 5.16PIDD201 pKa = 4.22SVCFGLSSRR210 pKa = 11.84VFYY213 pKa = 9.93PIHH216 pKa = 6.57RR217 pKa = 11.84RR218 pKa = 11.84LHH220 pKa = 5.64GRR222 pKa = 11.84VSLASRR228 pKa = 11.84RR229 pKa = 11.84NGPNTLDD236 pKa = 3.87GPNPARR242 pKa = 11.84PPPRR246 pKa = 11.84SPGLVDD252 pKa = 4.15RR253 pKa = 11.84EE254 pKa = 4.41AIRR257 pKa = 11.84LDD259 pKa = 3.26GQVYY263 pKa = 9.81CRR265 pKa = 11.84KK266 pKa = 9.95CGTSRR271 pKa = 11.84CEE273 pKa = 3.79LHH275 pKa = 5.71SHH277 pKa = 5.35LRR279 pKa = 11.84EE280 pKa = 3.76WMGPDD285 pKa = 3.39RR286 pKa = 11.84EE287 pKa = 4.38YY288 pKa = 11.33CSVSRR293 pKa = 11.84KK294 pKa = 9.24FGKK297 pKa = 9.81KK298 pKa = 9.24APAGARR304 pKa = 11.84EE305 pKa = 4.19SCYY308 pKa = 10.35KK309 pKa = 10.5RR310 pKa = 11.84SPKK313 pKa = 10.29NPSACGRR320 pKa = 11.84HH321 pKa = 4.71TPRR324 pKa = 11.84MRR326 pKa = 11.84ASAPVSPKK334 pKa = 10.35LQAVQMAA341 pKa = 4.15
Molecular weight: 37.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2870050 |
15 |
4924 |
546.3 |
60.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.808 ± 0.036 | 1.097 ± 0.01 |
5.706 ± 0.024 | 6.038 ± 0.034 |
3.605 ± 0.022 | 6.619 ± 0.029 |
2.317 ± 0.013 | 4.659 ± 0.024 |
4.901 ± 0.028 | 8.603 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.506 ± 0.014 | 3.631 ± 0.017 |
5.953 ± 0.037 | 3.911 ± 0.026 |
5.986 ± 0.026 | 8.393 ± 0.042 |
5.958 ± 0.025 | 6.393 ± 0.025 |
1.287 ± 0.011 | 2.63 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
