
Yangia sp. CCB-MM3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Salipiger; unclassified Salipiger
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
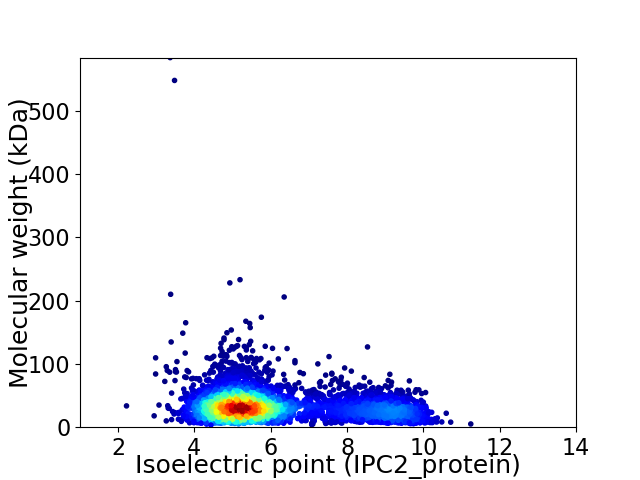
Virtual 2D-PAGE plot for 4896 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
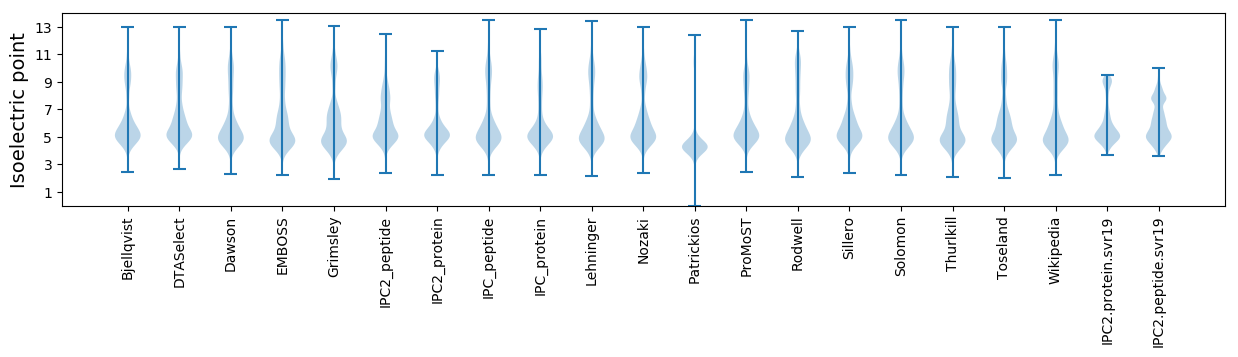
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B1PYL8|A0A1B1PYL8_9RHOB Uncharacterized protein OS=Yangia sp. CCB-MM3 OX=1792508 GN=AYJ57_24555 PE=4 SV=1
MM1 pKa = 7.53AADD4 pKa = 4.83FDD6 pKa = 4.6DD7 pKa = 6.06LDD9 pKa = 3.67VLAQQEE15 pKa = 4.28IAASEE20 pKa = 4.33SATNGNGANSLRR32 pKa = 11.84NLIEE36 pKa = 4.44RR37 pKa = 11.84GVLDD41 pKa = 3.61EE42 pKa = 4.44SGRR45 pKa = 11.84EE46 pKa = 3.88IGSEE50 pKa = 4.2SASPAPPGEE59 pKa = 4.51AGSGVLDD66 pKa = 3.48IFVEE70 pKa = 4.68DD71 pKa = 4.1GLLPGIRR78 pKa = 11.84SALEE82 pKa = 3.65QYY84 pKa = 10.61ISDD87 pKa = 4.31IEE89 pKa = 4.24LEE91 pKa = 4.69GYY93 pKa = 7.24QVRR96 pKa = 11.84VVEE99 pKa = 4.52FDD101 pKa = 3.42GSAEE105 pKa = 3.95EE106 pKa = 4.63LRR108 pKa = 11.84SEE110 pKa = 5.62LEE112 pKa = 4.01ARR114 pKa = 11.84WLNDD118 pKa = 3.12GLEE121 pKa = 4.36GALFVGDD128 pKa = 5.33LPVQYY133 pKa = 10.27FKK135 pKa = 11.28SYY137 pKa = 11.14DD138 pKa = 3.52GYY140 pKa = 11.31DD141 pKa = 3.13ADD143 pKa = 4.35GQLVRR148 pKa = 11.84YY149 pKa = 8.34PHH151 pKa = 7.16DD152 pKa = 4.61LYY154 pKa = 11.61FMDD157 pKa = 5.8LDD159 pKa = 3.91GSYY162 pKa = 11.13SFNASGADD170 pKa = 3.29AHH172 pKa = 6.95VDD174 pKa = 3.14GDD176 pKa = 4.01GDD178 pKa = 4.05VAPEE182 pKa = 4.06IYY184 pKa = 10.23VSRR187 pKa = 11.84LAASQLGGLSSSSEE201 pKa = 4.11TEE203 pKa = 4.34LINSYY208 pKa = 8.55FAKK211 pKa = 10.0LHH213 pKa = 5.15EE214 pKa = 4.25VRR216 pKa = 11.84AAGRR220 pKa = 11.84QYY222 pKa = 9.76QEE224 pKa = 3.53RR225 pKa = 11.84AIWFADD231 pKa = 3.77NDD233 pKa = 4.06WAPYY237 pKa = 9.57TGDD240 pKa = 3.97LEE242 pKa = 4.78SLTGLYY248 pKa = 10.43SEE250 pKa = 4.58VTDD253 pKa = 4.53IRR255 pKa = 11.84ALDD258 pKa = 3.69EE259 pKa = 4.31TTVANYY265 pKa = 7.79EE266 pKa = 3.84AALQEE271 pKa = 4.31DD272 pKa = 5.35VEE274 pKa = 4.47TLIQQIHH281 pKa = 6.0AWSSGLSISSPGGGLVFSADD301 pKa = 3.51VVDD304 pKa = 6.21LNAQPAFINLFNCSSADD321 pKa = 3.4FTQWSNLASTYY332 pKa = 10.88VFLSDD337 pKa = 4.36GVLNTVGSTKK347 pKa = 10.06TGSMLNFADD356 pKa = 5.76FYY358 pKa = 10.64RR359 pKa = 11.84PQSAGLTLGQSFLDD373 pKa = 3.65WFEE376 pKa = 3.78MHH378 pKa = 7.11AAATDD383 pKa = 3.77EE384 pKa = 4.79PALDD388 pKa = 3.8WKK390 pKa = 11.0VDD392 pKa = 2.9WHH394 pKa = 7.83YY395 pKa = 11.76GMTMQGDD402 pKa = 4.02PTMGPSVLRR411 pKa = 11.84AILPVITGTDD421 pKa = 3.22CRR423 pKa = 11.84DD424 pKa = 3.48RR425 pKa = 11.84LVGTDD430 pKa = 3.67DD431 pKa = 4.25AEE433 pKa = 5.08LILGLGGRR441 pKa = 11.84DD442 pKa = 3.81RR443 pKa = 11.84ITALAGDD450 pKa = 3.93DD451 pKa = 4.45RR452 pKa = 11.84IFGGAGRR459 pKa = 11.84DD460 pKa = 3.65TIRR463 pKa = 11.84AGDD466 pKa = 3.89GDD468 pKa = 4.38DD469 pKa = 5.52FIDD472 pKa = 4.25GGSGRR477 pKa = 11.84DD478 pKa = 3.54VIYY481 pKa = 10.17TGAGADD487 pKa = 3.42TVVISDD493 pKa = 4.01LRR495 pKa = 11.84GFDD498 pKa = 3.34KK499 pKa = 11.13LYY501 pKa = 11.04DD502 pKa = 3.71FDD504 pKa = 4.98IGVDD508 pKa = 3.46TLVIDD513 pKa = 4.46IEE515 pKa = 5.06GITPDD520 pKa = 4.97DD521 pKa = 3.81LSLNKK526 pKa = 8.57WRR528 pKa = 11.84LMASVDD534 pKa = 3.88GCWTKK539 pKa = 10.79LAFIPNAYY547 pKa = 9.99RR548 pKa = 11.84EE549 pKa = 4.37GATLAEE555 pKa = 4.32LLQPEE560 pKa = 4.7VPGDD564 pKa = 3.47ASTLYY569 pKa = 9.7TLDD572 pKa = 2.96VGSEE576 pKa = 3.98VRR578 pKa = 11.84GTLDD582 pKa = 3.62FLEE585 pKa = 4.49DD586 pKa = 3.79RR587 pKa = 11.84DD588 pKa = 3.86WFGVEE593 pKa = 3.53LRR595 pKa = 11.84AGVTYY600 pKa = 10.73RR601 pKa = 11.84FDD603 pKa = 3.71LVRR606 pKa = 11.84DD607 pKa = 3.82EE608 pKa = 5.37ASSNPLPDD616 pKa = 4.07PYY618 pKa = 10.82FRR620 pKa = 11.84LYY622 pKa = 10.67DD623 pKa = 3.85AEE625 pKa = 4.42GQLLAANDD633 pKa = 4.78DD634 pKa = 4.03YY635 pKa = 12.13SSLNSQLFYY644 pKa = 11.06TPTEE648 pKa = 4.04DD649 pKa = 4.01ARR651 pKa = 11.84VFAAAGAYY659 pKa = 10.01ADD661 pKa = 4.36AYY663 pKa = 9.63EE664 pKa = 4.44GGYY667 pKa = 9.97VLSLAEE673 pKa = 4.44AADD676 pKa = 4.09FDD678 pKa = 4.79AVPGDD683 pKa = 3.76EE684 pKa = 4.38STTAEE689 pKa = 4.19IEE691 pKa = 4.37VGGAAHH697 pKa = 6.93GRR699 pKa = 11.84IDD701 pKa = 4.76YY702 pKa = 10.39PGDD705 pKa = 4.03DD706 pKa = 3.2DD707 pKa = 3.79WFRR710 pKa = 11.84VEE712 pKa = 4.85LRR714 pKa = 11.84GGTTYY719 pKa = 10.95RR720 pKa = 11.84FYY722 pKa = 11.52LSAEE726 pKa = 4.3TGSDD730 pKa = 3.5SPLWDD735 pKa = 4.03PYY737 pKa = 10.97FYY739 pKa = 10.75LYY741 pKa = 10.64DD742 pKa = 4.25AEE744 pKa = 5.37GEE746 pKa = 4.31LIGANDD752 pKa = 3.82DD753 pKa = 4.05TGDD756 pKa = 4.16SLNSSYY762 pKa = 11.15QYY764 pKa = 10.86TPDD767 pKa = 3.23SDD769 pKa = 3.84EE770 pKa = 4.92LVFASARR777 pKa = 11.84AFGDD781 pKa = 3.67YY782 pKa = 9.42GTGDD786 pKa = 3.66YY787 pKa = 10.71VLSVAEE793 pKa = 4.5KK794 pKa = 9.57EE795 pKa = 4.08LGLFGMLQDD804 pKa = 3.54VPAYY808 pKa = 9.75NWYY811 pKa = 9.74HH812 pKa = 5.85GCSPTTGGSIIGYY825 pKa = 8.56WDD827 pKa = 3.75LNGYY831 pKa = 8.92PDD833 pKa = 5.23LFDD836 pKa = 4.81AEE838 pKa = 4.4GWEE841 pKa = 4.46QISTTPSVQDD851 pKa = 3.69QISSPAHH858 pKa = 4.97NAKK861 pKa = 10.24YY862 pKa = 10.59DD863 pKa = 3.8PTPDD867 pKa = 3.67NPTLPVPPYY876 pKa = 10.11TSIADD881 pKa = 3.67FMGTSVDD888 pKa = 3.49PLGYY892 pKa = 10.41GSTYY896 pKa = 10.57VSNFDD901 pKa = 3.08GAYY904 pKa = 9.62RR905 pKa = 11.84GYY907 pKa = 9.9PALTGSAFSSHH918 pKa = 5.63TLYY921 pKa = 11.16NSDD924 pKa = 3.13TGEE927 pKa = 4.3PGGSLWEE934 pKa = 4.25EE935 pKa = 4.03YY936 pKa = 10.34VAEE939 pKa = 4.34IDD941 pKa = 3.25AGRR944 pKa = 11.84PVAMTVDD951 pKa = 3.87SNGDD955 pKa = 3.67GITDD959 pKa = 3.52HH960 pKa = 7.07SIPGIGYY967 pKa = 8.19EE968 pKa = 4.56DD969 pKa = 4.04RR970 pKa = 11.84GADD973 pKa = 3.91GVWYY977 pKa = 9.68AAYY980 pKa = 6.51TTWSEE985 pKa = 4.22SEE987 pKa = 4.08TPIWRR992 pKa = 11.84EE993 pKa = 3.7FRR995 pKa = 11.84PTAPGAAWAVSYY1007 pKa = 8.71LTFVLPEE1014 pKa = 4.09GAEE1017 pKa = 3.92AAPTGDD1023 pKa = 4.4LALAMAGGAEE1033 pKa = 4.84PITSEE1038 pKa = 3.91PLTFTEE1044 pKa = 4.49EE1045 pKa = 3.76LHH1047 pKa = 6.63EE1048 pKa = 4.2IAVALEE1054 pKa = 3.69IDD1056 pKa = 4.14GEE1058 pKa = 4.27LGAAEE1063 pKa = 4.73EE1064 pKa = 4.75KK1065 pKa = 10.66ILMQAKK1071 pKa = 10.27LIDD1074 pKa = 3.7FAQPFDD1080 pKa = 4.03DD1081 pKa = 4.87LFVVV1085 pKa = 4.13
MM1 pKa = 7.53AADD4 pKa = 4.83FDD6 pKa = 4.6DD7 pKa = 6.06LDD9 pKa = 3.67VLAQQEE15 pKa = 4.28IAASEE20 pKa = 4.33SATNGNGANSLRR32 pKa = 11.84NLIEE36 pKa = 4.44RR37 pKa = 11.84GVLDD41 pKa = 3.61EE42 pKa = 4.44SGRR45 pKa = 11.84EE46 pKa = 3.88IGSEE50 pKa = 4.2SASPAPPGEE59 pKa = 4.51AGSGVLDD66 pKa = 3.48IFVEE70 pKa = 4.68DD71 pKa = 4.1GLLPGIRR78 pKa = 11.84SALEE82 pKa = 3.65QYY84 pKa = 10.61ISDD87 pKa = 4.31IEE89 pKa = 4.24LEE91 pKa = 4.69GYY93 pKa = 7.24QVRR96 pKa = 11.84VVEE99 pKa = 4.52FDD101 pKa = 3.42GSAEE105 pKa = 3.95EE106 pKa = 4.63LRR108 pKa = 11.84SEE110 pKa = 5.62LEE112 pKa = 4.01ARR114 pKa = 11.84WLNDD118 pKa = 3.12GLEE121 pKa = 4.36GALFVGDD128 pKa = 5.33LPVQYY133 pKa = 10.27FKK135 pKa = 11.28SYY137 pKa = 11.14DD138 pKa = 3.52GYY140 pKa = 11.31DD141 pKa = 3.13ADD143 pKa = 4.35GQLVRR148 pKa = 11.84YY149 pKa = 8.34PHH151 pKa = 7.16DD152 pKa = 4.61LYY154 pKa = 11.61FMDD157 pKa = 5.8LDD159 pKa = 3.91GSYY162 pKa = 11.13SFNASGADD170 pKa = 3.29AHH172 pKa = 6.95VDD174 pKa = 3.14GDD176 pKa = 4.01GDD178 pKa = 4.05VAPEE182 pKa = 4.06IYY184 pKa = 10.23VSRR187 pKa = 11.84LAASQLGGLSSSSEE201 pKa = 4.11TEE203 pKa = 4.34LINSYY208 pKa = 8.55FAKK211 pKa = 10.0LHH213 pKa = 5.15EE214 pKa = 4.25VRR216 pKa = 11.84AAGRR220 pKa = 11.84QYY222 pKa = 9.76QEE224 pKa = 3.53RR225 pKa = 11.84AIWFADD231 pKa = 3.77NDD233 pKa = 4.06WAPYY237 pKa = 9.57TGDD240 pKa = 3.97LEE242 pKa = 4.78SLTGLYY248 pKa = 10.43SEE250 pKa = 4.58VTDD253 pKa = 4.53IRR255 pKa = 11.84ALDD258 pKa = 3.69EE259 pKa = 4.31TTVANYY265 pKa = 7.79EE266 pKa = 3.84AALQEE271 pKa = 4.31DD272 pKa = 5.35VEE274 pKa = 4.47TLIQQIHH281 pKa = 6.0AWSSGLSISSPGGGLVFSADD301 pKa = 3.51VVDD304 pKa = 6.21LNAQPAFINLFNCSSADD321 pKa = 3.4FTQWSNLASTYY332 pKa = 10.88VFLSDD337 pKa = 4.36GVLNTVGSTKK347 pKa = 10.06TGSMLNFADD356 pKa = 5.76FYY358 pKa = 10.64RR359 pKa = 11.84PQSAGLTLGQSFLDD373 pKa = 3.65WFEE376 pKa = 3.78MHH378 pKa = 7.11AAATDD383 pKa = 3.77EE384 pKa = 4.79PALDD388 pKa = 3.8WKK390 pKa = 11.0VDD392 pKa = 2.9WHH394 pKa = 7.83YY395 pKa = 11.76GMTMQGDD402 pKa = 4.02PTMGPSVLRR411 pKa = 11.84AILPVITGTDD421 pKa = 3.22CRR423 pKa = 11.84DD424 pKa = 3.48RR425 pKa = 11.84LVGTDD430 pKa = 3.67DD431 pKa = 4.25AEE433 pKa = 5.08LILGLGGRR441 pKa = 11.84DD442 pKa = 3.81RR443 pKa = 11.84ITALAGDD450 pKa = 3.93DD451 pKa = 4.45RR452 pKa = 11.84IFGGAGRR459 pKa = 11.84DD460 pKa = 3.65TIRR463 pKa = 11.84AGDD466 pKa = 3.89GDD468 pKa = 4.38DD469 pKa = 5.52FIDD472 pKa = 4.25GGSGRR477 pKa = 11.84DD478 pKa = 3.54VIYY481 pKa = 10.17TGAGADD487 pKa = 3.42TVVISDD493 pKa = 4.01LRR495 pKa = 11.84GFDD498 pKa = 3.34KK499 pKa = 11.13LYY501 pKa = 11.04DD502 pKa = 3.71FDD504 pKa = 4.98IGVDD508 pKa = 3.46TLVIDD513 pKa = 4.46IEE515 pKa = 5.06GITPDD520 pKa = 4.97DD521 pKa = 3.81LSLNKK526 pKa = 8.57WRR528 pKa = 11.84LMASVDD534 pKa = 3.88GCWTKK539 pKa = 10.79LAFIPNAYY547 pKa = 9.99RR548 pKa = 11.84EE549 pKa = 4.37GATLAEE555 pKa = 4.32LLQPEE560 pKa = 4.7VPGDD564 pKa = 3.47ASTLYY569 pKa = 9.7TLDD572 pKa = 2.96VGSEE576 pKa = 3.98VRR578 pKa = 11.84GTLDD582 pKa = 3.62FLEE585 pKa = 4.49DD586 pKa = 3.79RR587 pKa = 11.84DD588 pKa = 3.86WFGVEE593 pKa = 3.53LRR595 pKa = 11.84AGVTYY600 pKa = 10.73RR601 pKa = 11.84FDD603 pKa = 3.71LVRR606 pKa = 11.84DD607 pKa = 3.82EE608 pKa = 5.37ASSNPLPDD616 pKa = 4.07PYY618 pKa = 10.82FRR620 pKa = 11.84LYY622 pKa = 10.67DD623 pKa = 3.85AEE625 pKa = 4.42GQLLAANDD633 pKa = 4.78DD634 pKa = 4.03YY635 pKa = 12.13SSLNSQLFYY644 pKa = 11.06TPTEE648 pKa = 4.04DD649 pKa = 4.01ARR651 pKa = 11.84VFAAAGAYY659 pKa = 10.01ADD661 pKa = 4.36AYY663 pKa = 9.63EE664 pKa = 4.44GGYY667 pKa = 9.97VLSLAEE673 pKa = 4.44AADD676 pKa = 4.09FDD678 pKa = 4.79AVPGDD683 pKa = 3.76EE684 pKa = 4.38STTAEE689 pKa = 4.19IEE691 pKa = 4.37VGGAAHH697 pKa = 6.93GRR699 pKa = 11.84IDD701 pKa = 4.76YY702 pKa = 10.39PGDD705 pKa = 4.03DD706 pKa = 3.2DD707 pKa = 3.79WFRR710 pKa = 11.84VEE712 pKa = 4.85LRR714 pKa = 11.84GGTTYY719 pKa = 10.95RR720 pKa = 11.84FYY722 pKa = 11.52LSAEE726 pKa = 4.3TGSDD730 pKa = 3.5SPLWDD735 pKa = 4.03PYY737 pKa = 10.97FYY739 pKa = 10.75LYY741 pKa = 10.64DD742 pKa = 4.25AEE744 pKa = 5.37GEE746 pKa = 4.31LIGANDD752 pKa = 3.82DD753 pKa = 4.05TGDD756 pKa = 4.16SLNSSYY762 pKa = 11.15QYY764 pKa = 10.86TPDD767 pKa = 3.23SDD769 pKa = 3.84EE770 pKa = 4.92LVFASARR777 pKa = 11.84AFGDD781 pKa = 3.67YY782 pKa = 9.42GTGDD786 pKa = 3.66YY787 pKa = 10.71VLSVAEE793 pKa = 4.5KK794 pKa = 9.57EE795 pKa = 4.08LGLFGMLQDD804 pKa = 3.54VPAYY808 pKa = 9.75NWYY811 pKa = 9.74HH812 pKa = 5.85GCSPTTGGSIIGYY825 pKa = 8.56WDD827 pKa = 3.75LNGYY831 pKa = 8.92PDD833 pKa = 5.23LFDD836 pKa = 4.81AEE838 pKa = 4.4GWEE841 pKa = 4.46QISTTPSVQDD851 pKa = 3.69QISSPAHH858 pKa = 4.97NAKK861 pKa = 10.24YY862 pKa = 10.59DD863 pKa = 3.8PTPDD867 pKa = 3.67NPTLPVPPYY876 pKa = 10.11TSIADD881 pKa = 3.67FMGTSVDD888 pKa = 3.49PLGYY892 pKa = 10.41GSTYY896 pKa = 10.57VSNFDD901 pKa = 3.08GAYY904 pKa = 9.62RR905 pKa = 11.84GYY907 pKa = 9.9PALTGSAFSSHH918 pKa = 5.63TLYY921 pKa = 11.16NSDD924 pKa = 3.13TGEE927 pKa = 4.3PGGSLWEE934 pKa = 4.25EE935 pKa = 4.03YY936 pKa = 10.34VAEE939 pKa = 4.34IDD941 pKa = 3.25AGRR944 pKa = 11.84PVAMTVDD951 pKa = 3.87SNGDD955 pKa = 3.67GITDD959 pKa = 3.52HH960 pKa = 7.07SIPGIGYY967 pKa = 8.19EE968 pKa = 4.56DD969 pKa = 4.04RR970 pKa = 11.84GADD973 pKa = 3.91GVWYY977 pKa = 9.68AAYY980 pKa = 6.51TTWSEE985 pKa = 4.22SEE987 pKa = 4.08TPIWRR992 pKa = 11.84EE993 pKa = 3.7FRR995 pKa = 11.84PTAPGAAWAVSYY1007 pKa = 8.71LTFVLPEE1014 pKa = 4.09GAEE1017 pKa = 3.92AAPTGDD1023 pKa = 4.4LALAMAGGAEE1033 pKa = 4.84PITSEE1038 pKa = 3.91PLTFTEE1044 pKa = 4.49EE1045 pKa = 3.76LHH1047 pKa = 6.63EE1048 pKa = 4.2IAVALEE1054 pKa = 3.69IDD1056 pKa = 4.14GEE1058 pKa = 4.27LGAAEE1063 pKa = 4.73EE1064 pKa = 4.75KK1065 pKa = 10.66ILMQAKK1071 pKa = 10.27LIDD1074 pKa = 3.7FAQPFDD1080 pKa = 4.03DD1081 pKa = 4.87LFVVV1085 pKa = 4.13
Molecular weight: 117.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B1PN51|A0A1B1PN51_9RHOB Uncharacterized protein OS=Yangia sp. CCB-MM3 OX=1792508 GN=AYJ57_06330 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.31AGRR28 pKa = 11.84AILNARR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06KK41 pKa = 10.57LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.31AGRR28 pKa = 11.84AILNARR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06KK41 pKa = 10.57LSAA44 pKa = 4.03
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1570501 |
30 |
5754 |
320.8 |
34.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.588 ± 0.043 | 0.901 ± 0.012 |
5.742 ± 0.04 | 6.359 ± 0.037 |
3.67 ± 0.024 | 8.914 ± 0.034 |
1.991 ± 0.02 | 4.881 ± 0.027 |
2.941 ± 0.029 | 10.479 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.768 ± 0.02 | 2.333 ± 0.019 |
5.237 ± 0.025 | 3.182 ± 0.019 |
6.657 ± 0.046 | 5.303 ± 0.031 |
5.307 ± 0.031 | 7.142 ± 0.032 |
1.388 ± 0.014 | 2.215 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
