
Alistipes sp. An31A
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; unclassified Alistipes
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
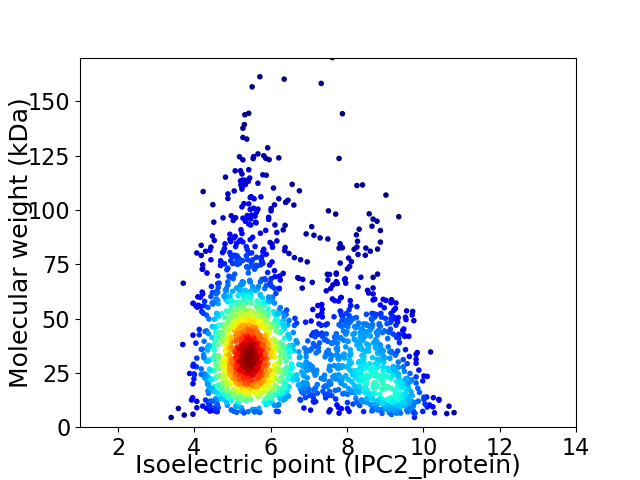
Virtual 2D-PAGE plot for 2054 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4ARL2|A0A1Y4ARL2_9BACT Uncharacterized protein OS=Alistipes sp. An31A OX=1965631 GN=B5F90_05205 PE=4 SV=1
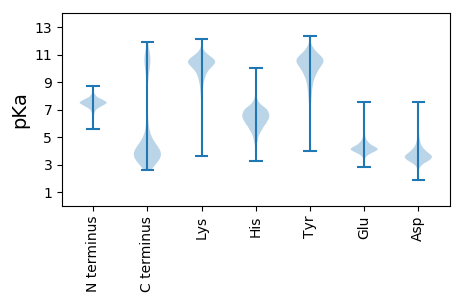
MM1 pKa = 7.04KK2 pKa = 10.22QLYY5 pKa = 9.75LLPLLAILALTGCKK19 pKa = 9.65KK20 pKa = 10.72DD21 pKa = 4.46DD22 pKa = 4.17DD23 pKa = 4.46ASRR26 pKa = 11.84PGNLTIEE33 pKa = 4.27KK34 pKa = 9.64TEE36 pKa = 3.95YY37 pKa = 10.48TLDD40 pKa = 3.64ADD42 pKa = 4.39EE43 pKa = 4.95EE44 pKa = 4.43RR45 pKa = 11.84TATVAFTAAADD56 pKa = 3.5WTLSVVYY63 pKa = 10.53DD64 pKa = 4.19DD65 pKa = 5.58AEE67 pKa = 4.51SSADD71 pKa = 3.35WLTVTPTSGTAGEE84 pKa = 4.26QTVEE88 pKa = 4.36LSATRR93 pKa = 11.84NFRR96 pKa = 11.84ASARR100 pKa = 11.84TAYY103 pKa = 10.57ADD105 pKa = 4.14LACGEE110 pKa = 4.08QSVRR114 pKa = 11.84LTVTQTAASGDD125 pKa = 3.68TDD127 pKa = 3.89FTAQFDD133 pKa = 4.01PDD135 pKa = 3.78FAKK138 pKa = 10.45EE139 pKa = 3.93LQKK142 pKa = 10.92QGIIADD148 pKa = 4.07AGNITPEE155 pKa = 4.1DD156 pKa = 3.79MEE158 pKa = 5.42KK159 pKa = 10.2IAAITSLDD167 pKa = 3.48VSGTEE172 pKa = 4.25DD173 pKa = 3.53APGALTSLQGIEE185 pKa = 4.16YY186 pKa = 10.2FEE188 pKa = 4.72SLTEE192 pKa = 3.94LEE194 pKa = 4.26CRR196 pKa = 11.84YY197 pKa = 10.14NQLTSLDD204 pKa = 3.66VSRR207 pKa = 11.84NTALMEE213 pKa = 4.85LEE215 pKa = 4.16CQYY218 pKa = 11.47NQLPEE223 pKa = 4.97LDD225 pKa = 3.55VSANTKK231 pKa = 9.87LWLLICSSNSMEE243 pKa = 4.19TLNVGANTALTYY255 pKa = 9.61LYY257 pKa = 10.73CNYY260 pKa = 10.57NEE262 pKa = 4.35LTTLDD267 pKa = 3.4VSRR270 pKa = 11.84NTEE273 pKa = 4.13LMYY276 pKa = 10.64LSCFNNKK283 pKa = 8.01LTEE286 pKa = 4.23LDD288 pKa = 3.28VRR290 pKa = 11.84ANTKK294 pKa = 10.21LADD297 pKa = 4.28LSCSNNSLTEE307 pKa = 3.76LDD309 pKa = 3.43VRR311 pKa = 11.84ANTEE315 pKa = 4.35LEE317 pKa = 4.29MFDD320 pKa = 4.58CSNNSLTEE328 pKa = 3.96LDD330 pKa = 3.59VSANTKK336 pKa = 9.72LAWHH340 pKa = 6.96FYY342 pKa = 10.66CYY344 pKa = 10.79GNQLTEE350 pKa = 4.55LDD352 pKa = 4.05VSNCTALEE360 pKa = 3.91VLFCHH365 pKa = 7.07DD366 pKa = 4.66NPLTSLDD373 pKa = 3.62LSRR376 pKa = 11.84NTALTEE382 pKa = 4.1LQCSNNQLTEE392 pKa = 4.11LDD394 pKa = 3.75LSRR397 pKa = 11.84NTALTKK403 pKa = 10.33LRR405 pKa = 11.84CFDD408 pKa = 4.21NEE410 pKa = 4.31LTSLDD415 pKa = 3.61LSRR418 pKa = 11.84NTVLTDD424 pKa = 3.69LYY426 pKa = 10.91CGNNQLTSLDD436 pKa = 3.65VSQNTALTEE445 pKa = 3.89LSCFSNEE452 pKa = 4.52LPSLDD457 pKa = 4.02LSRR460 pKa = 11.84NTALTLLDD468 pKa = 3.8CSYY471 pKa = 11.51NPGDD475 pKa = 3.99GVSSFPVTAWFDD487 pKa = 3.81NEE489 pKa = 4.38TVPAGLDD496 pKa = 3.59VYY498 pKa = 11.22SSSWWYY504 pKa = 10.12DD505 pKa = 3.27DD506 pKa = 4.25KK507 pKa = 11.82NITIDD512 pKa = 3.83FRR514 pKa = 11.84KK515 pKa = 8.68TEE517 pKa = 3.83
MM1 pKa = 7.04KK2 pKa = 10.22QLYY5 pKa = 9.75LLPLLAILALTGCKK19 pKa = 9.65KK20 pKa = 10.72DD21 pKa = 4.46DD22 pKa = 4.17DD23 pKa = 4.46ASRR26 pKa = 11.84PGNLTIEE33 pKa = 4.27KK34 pKa = 9.64TEE36 pKa = 3.95YY37 pKa = 10.48TLDD40 pKa = 3.64ADD42 pKa = 4.39EE43 pKa = 4.95EE44 pKa = 4.43RR45 pKa = 11.84TATVAFTAAADD56 pKa = 3.5WTLSVVYY63 pKa = 10.53DD64 pKa = 4.19DD65 pKa = 5.58AEE67 pKa = 4.51SSADD71 pKa = 3.35WLTVTPTSGTAGEE84 pKa = 4.26QTVEE88 pKa = 4.36LSATRR93 pKa = 11.84NFRR96 pKa = 11.84ASARR100 pKa = 11.84TAYY103 pKa = 10.57ADD105 pKa = 4.14LACGEE110 pKa = 4.08QSVRR114 pKa = 11.84LTVTQTAASGDD125 pKa = 3.68TDD127 pKa = 3.89FTAQFDD133 pKa = 4.01PDD135 pKa = 3.78FAKK138 pKa = 10.45EE139 pKa = 3.93LQKK142 pKa = 10.92QGIIADD148 pKa = 4.07AGNITPEE155 pKa = 4.1DD156 pKa = 3.79MEE158 pKa = 5.42KK159 pKa = 10.2IAAITSLDD167 pKa = 3.48VSGTEE172 pKa = 4.25DD173 pKa = 3.53APGALTSLQGIEE185 pKa = 4.16YY186 pKa = 10.2FEE188 pKa = 4.72SLTEE192 pKa = 3.94LEE194 pKa = 4.26CRR196 pKa = 11.84YY197 pKa = 10.14NQLTSLDD204 pKa = 3.66VSRR207 pKa = 11.84NTALMEE213 pKa = 4.85LEE215 pKa = 4.16CQYY218 pKa = 11.47NQLPEE223 pKa = 4.97LDD225 pKa = 3.55VSANTKK231 pKa = 9.87LWLLICSSNSMEE243 pKa = 4.19TLNVGANTALTYY255 pKa = 9.61LYY257 pKa = 10.73CNYY260 pKa = 10.57NEE262 pKa = 4.35LTTLDD267 pKa = 3.4VSRR270 pKa = 11.84NTEE273 pKa = 4.13LMYY276 pKa = 10.64LSCFNNKK283 pKa = 8.01LTEE286 pKa = 4.23LDD288 pKa = 3.28VRR290 pKa = 11.84ANTKK294 pKa = 10.21LADD297 pKa = 4.28LSCSNNSLTEE307 pKa = 3.76LDD309 pKa = 3.43VRR311 pKa = 11.84ANTEE315 pKa = 4.35LEE317 pKa = 4.29MFDD320 pKa = 4.58CSNNSLTEE328 pKa = 3.96LDD330 pKa = 3.59VSANTKK336 pKa = 9.72LAWHH340 pKa = 6.96FYY342 pKa = 10.66CYY344 pKa = 10.79GNQLTEE350 pKa = 4.55LDD352 pKa = 4.05VSNCTALEE360 pKa = 3.91VLFCHH365 pKa = 7.07DD366 pKa = 4.66NPLTSLDD373 pKa = 3.62LSRR376 pKa = 11.84NTALTEE382 pKa = 4.1LQCSNNQLTEE392 pKa = 4.11LDD394 pKa = 3.75LSRR397 pKa = 11.84NTALTKK403 pKa = 10.33LRR405 pKa = 11.84CFDD408 pKa = 4.21NEE410 pKa = 4.31LTSLDD415 pKa = 3.61LSRR418 pKa = 11.84NTVLTDD424 pKa = 3.69LYY426 pKa = 10.91CGNNQLTSLDD436 pKa = 3.65VSQNTALTEE445 pKa = 3.89LSCFSNEE452 pKa = 4.52LPSLDD457 pKa = 4.02LSRR460 pKa = 11.84NTALTLLDD468 pKa = 3.8CSYY471 pKa = 11.51NPGDD475 pKa = 3.99GVSSFPVTAWFDD487 pKa = 3.81NEE489 pKa = 4.38TVPAGLDD496 pKa = 3.59VYY498 pKa = 11.22SSSWWYY504 pKa = 10.12DD505 pKa = 3.27DD506 pKa = 4.25KK507 pKa = 11.82NITIDD512 pKa = 3.83FRR514 pKa = 11.84KK515 pKa = 8.68TEE517 pKa = 3.83
Molecular weight: 57.01 kDa
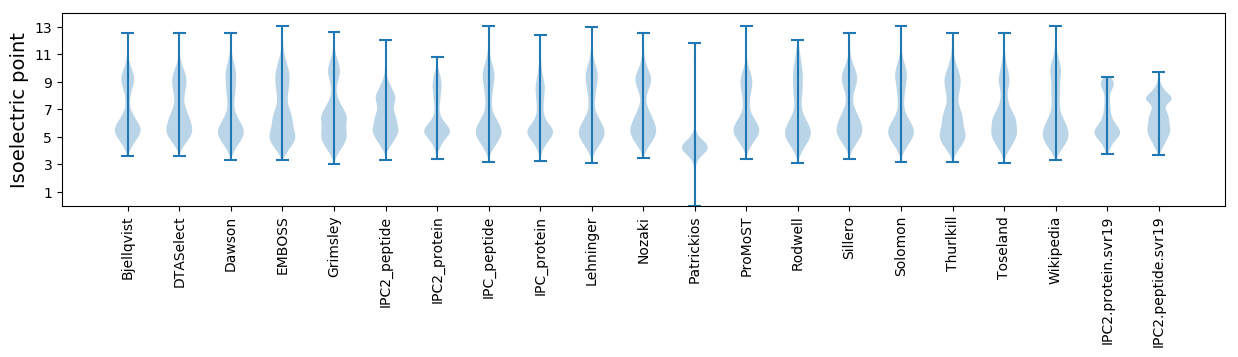
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4AE02|A0A1Y4AE02_9BACT Mannosyltransferase OS=Alistipes sp. An31A OX=1965631 GN=B5F90_06675 PE=4 SV=1
MM1 pKa = 7.49PSSRR5 pKa = 11.84RR6 pKa = 11.84GPCGCRR12 pKa = 11.84ASAAPRR18 pKa = 11.84GGRR21 pKa = 11.84RR22 pKa = 11.84PPAPGRR28 pKa = 11.84RR29 pKa = 11.84SRR31 pKa = 11.84RR32 pKa = 11.84AAGGSPPRR40 pKa = 11.84GAGRR44 pKa = 11.84RR45 pKa = 11.84GGVRR49 pKa = 11.84PPGAGSAPRR58 pKa = 11.84RR59 pKa = 11.84PRR61 pKa = 11.84CRR63 pKa = 11.84PRR65 pKa = 4.6
MM1 pKa = 7.49PSSRR5 pKa = 11.84RR6 pKa = 11.84GPCGCRR12 pKa = 11.84ASAAPRR18 pKa = 11.84GGRR21 pKa = 11.84RR22 pKa = 11.84PPAPGRR28 pKa = 11.84RR29 pKa = 11.84SRR31 pKa = 11.84RR32 pKa = 11.84AAGGSPPRR40 pKa = 11.84GAGRR44 pKa = 11.84RR45 pKa = 11.84GGVRR49 pKa = 11.84PPGAGSAPRR58 pKa = 11.84RR59 pKa = 11.84PRR61 pKa = 11.84CRR63 pKa = 11.84PRR65 pKa = 4.6
Molecular weight: 6.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
728213 |
38 |
1533 |
354.5 |
39.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.492 ± 0.063 | 1.305 ± 0.021 |
5.419 ± 0.036 | 7.055 ± 0.052 |
4.122 ± 0.032 | 7.51 ± 0.044 |
1.906 ± 0.021 | 5.406 ± 0.046 |
4.063 ± 0.053 | 9.743 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.544 ± 0.025 | 3.542 ± 0.037 |
4.223 ± 0.033 | 3.12 ± 0.029 |
7.157 ± 0.061 | 5.743 ± 0.039 |
5.617 ± 0.046 | 6.981 ± 0.044 |
1.223 ± 0.021 | 3.829 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
