
Delphinapterus leucas (Beluga whale)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria; Artiodactyla;
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
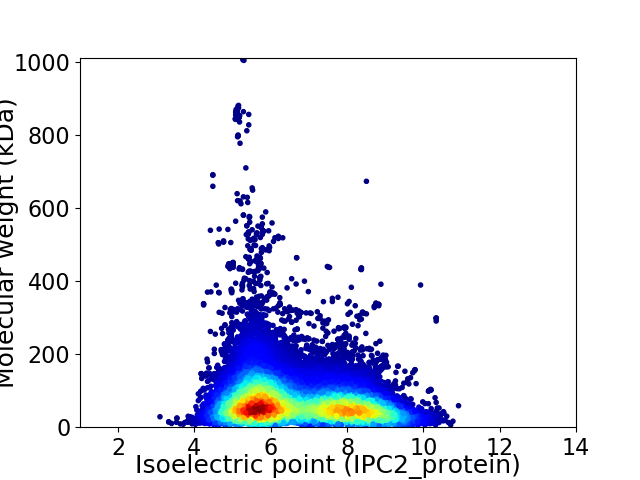
Virtual 2D-PAGE plot for 40783 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Y9PN18|A0A2Y9PN18_DELLE Isoform of A0A2Y9PCP3 transmembrane and coiled-coil domain-containing protein 6 isoform X4 OS=Delphinapterus leucas OX=9749 GN=TMCO6 PE=4 SV=1
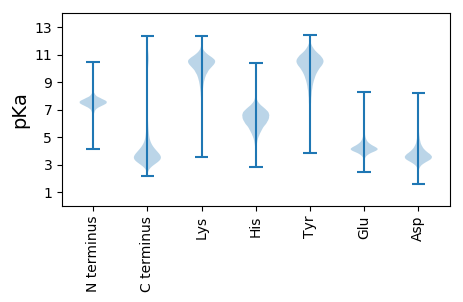
MM1 pKa = 7.3EE2 pKa = 5.17AVSLVLFVLGLLLLPLLAVLLMALCVRR29 pKa = 11.84CRR31 pKa = 11.84EE32 pKa = 4.25LPGSYY37 pKa = 8.67DD38 pKa = 3.24TAASDD43 pKa = 3.91GLTPSSIVIKK53 pKa = 9.83PPPTVAPWPPATSYY67 pKa = 11.3PPVTYY72 pKa = 10.68LLQSQPDD79 pKa = 3.91LLRR82 pKa = 11.84IPRR85 pKa = 11.84SPQPPGGSPRR95 pKa = 11.84MPSSQQDD102 pKa = 3.01SDD104 pKa = 3.88GANSVASYY112 pKa = 10.42EE113 pKa = 4.12NEE115 pKa = 3.82EE116 pKa = 4.38AVCEE120 pKa = 4.31DD121 pKa = 3.54ADD123 pKa = 3.94EE124 pKa = 6.05DD125 pKa = 4.37EE126 pKa = 5.92DD127 pKa = 4.86EE128 pKa = 4.15EE129 pKa = 5.22DD130 pKa = 3.81YY131 pKa = 11.56PEE133 pKa = 5.32GYY135 pKa = 10.61LVVLPDD141 pKa = 3.71NVPATGAAVPPAPASSNPGLRR162 pKa = 11.84DD163 pKa = 3.06SAFSMEE169 pKa = 4.23SGEE172 pKa = 4.99DD173 pKa = 3.63YY174 pKa = 11.66VNVPEE179 pKa = 4.96SEE181 pKa = 4.26GSADD185 pKa = 3.47ASLDD189 pKa = 3.39GSRR192 pKa = 11.84EE193 pKa = 3.99YY194 pKa = 11.72VNVSQEE200 pKa = 4.24LPPVARR206 pKa = 11.84TKK208 pKa = 10.27PAGRR212 pKa = 11.84SSQEE216 pKa = 3.88VEE218 pKa = 4.13GEE220 pKa = 3.96EE221 pKa = 4.31APDD224 pKa = 3.64YY225 pKa = 11.32EE226 pKa = 4.44NLQIHH231 pKa = 6.6
MM1 pKa = 7.3EE2 pKa = 5.17AVSLVLFVLGLLLLPLLAVLLMALCVRR29 pKa = 11.84CRR31 pKa = 11.84EE32 pKa = 4.25LPGSYY37 pKa = 8.67DD38 pKa = 3.24TAASDD43 pKa = 3.91GLTPSSIVIKK53 pKa = 9.83PPPTVAPWPPATSYY67 pKa = 11.3PPVTYY72 pKa = 10.68LLQSQPDD79 pKa = 3.91LLRR82 pKa = 11.84IPRR85 pKa = 11.84SPQPPGGSPRR95 pKa = 11.84MPSSQQDD102 pKa = 3.01SDD104 pKa = 3.88GANSVASYY112 pKa = 10.42EE113 pKa = 4.12NEE115 pKa = 3.82EE116 pKa = 4.38AVCEE120 pKa = 4.31DD121 pKa = 3.54ADD123 pKa = 3.94EE124 pKa = 6.05DD125 pKa = 4.37EE126 pKa = 5.92DD127 pKa = 4.86EE128 pKa = 4.15EE129 pKa = 5.22DD130 pKa = 3.81YY131 pKa = 11.56PEE133 pKa = 5.32GYY135 pKa = 10.61LVVLPDD141 pKa = 3.71NVPATGAAVPPAPASSNPGLRR162 pKa = 11.84DD163 pKa = 3.06SAFSMEE169 pKa = 4.23SGEE172 pKa = 4.99DD173 pKa = 3.63YY174 pKa = 11.66VNVPEE179 pKa = 4.96SEE181 pKa = 4.26GSADD185 pKa = 3.47ASLDD189 pKa = 3.39GSRR192 pKa = 11.84EE193 pKa = 3.99YY194 pKa = 11.72VNVSQEE200 pKa = 4.24LPPVARR206 pKa = 11.84TKK208 pKa = 10.27PAGRR212 pKa = 11.84SSQEE216 pKa = 3.88VEE218 pKa = 4.13GEE220 pKa = 3.96EE221 pKa = 4.31APDD224 pKa = 3.64YY225 pKa = 11.32EE226 pKa = 4.44NLQIHH231 pKa = 6.6
Molecular weight: 24.51 kDa
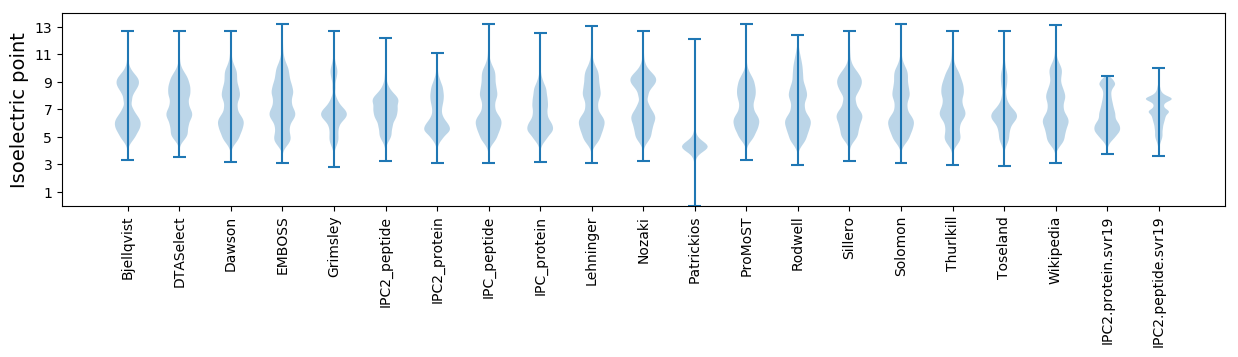
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Y9M5L6|A0A2Y9M5L6_DELLE Phosphodiesterase OS=Delphinapterus leucas OX=9749 GN=PDE4C PE=3 SV=1
MM1 pKa = 7.18GWAALAGPARR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84VLQTARR20 pKa = 11.84RR21 pKa = 11.84ARR23 pKa = 11.84QTTLSRR29 pKa = 11.84PGPPDD34 pKa = 3.06QTALAGPHH42 pKa = 3.99QTRR45 pKa = 11.84PRR47 pKa = 11.84RR48 pKa = 11.84VLQAALSRR56 pKa = 11.84PGPPDD61 pKa = 2.82QAARR65 pKa = 11.84PRR67 pKa = 11.84PVRR70 pKa = 11.84PRR72 pKa = 11.84SPGRR76 pKa = 11.84ARR78 pKa = 11.84QTRR81 pKa = 11.84PRR83 pKa = 11.84SPRR86 pKa = 11.84PARR89 pKa = 11.84PRR91 pKa = 11.84RR92 pKa = 11.84VLQTALSRR100 pKa = 11.84PGPPDD105 pKa = 3.0RR106 pKa = 11.84AGSSRR111 pKa = 11.84PRR113 pKa = 11.84AGPARR118 pKa = 11.84PRR120 pKa = 11.84AGSSRR125 pKa = 11.84PRR127 pKa = 11.84AGPAGPRR134 pKa = 11.84QAHH137 pKa = 3.87QTARR141 pKa = 11.84ARR143 pKa = 11.84SSRR146 pKa = 11.84PRR148 pKa = 11.84SPGRR152 pKa = 11.84VRR154 pKa = 11.84QTRR157 pKa = 11.84PRR159 pKa = 11.84SLGPPDD165 pKa = 3.69QPEE168 pKa = 4.02LAGPARR174 pKa = 11.84SRR176 pKa = 11.84RR177 pKa = 11.84VLRR180 pKa = 11.84TAPGRR185 pKa = 11.84ARR187 pKa = 11.84QVLQTTVSRR196 pKa = 11.84PGPPNQATRR205 pKa = 11.84AGPTRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84SPGRR216 pKa = 11.84VRR218 pKa = 11.84QTRR221 pKa = 11.84PSSPSQAAPGPSDD234 pKa = 3.35PAPRR238 pKa = 11.84ARR240 pKa = 11.84QAALTRR246 pKa = 11.84ARR248 pKa = 11.84QAAPGHH254 pKa = 6.84PDD256 pKa = 3.06RR257 pKa = 11.84ALQAGSARR265 pKa = 11.84PGRR268 pKa = 11.84ARR270 pKa = 11.84PGPPDD275 pKa = 3.2QTALARR281 pKa = 11.84PQQTRR286 pKa = 11.84PRR288 pKa = 11.84RR289 pKa = 11.84VLQAALSRR297 pKa = 11.84PGPPDD302 pKa = 3.18QATHH306 pKa = 7.16PGPARR311 pKa = 11.84PARR314 pKa = 11.84PRR316 pKa = 11.84PPGRR320 pKa = 11.84APQTGPRR327 pKa = 11.84SPAPARR333 pKa = 11.84PNRR336 pKa = 11.84TCRR339 pKa = 11.84ARR341 pKa = 11.84QAVAGPPDD349 pKa = 3.9RR350 pKa = 11.84GLQAGATRR358 pKa = 11.84PRR360 pKa = 11.84RR361 pKa = 11.84VRR363 pKa = 11.84QTTRR367 pKa = 11.84WALQTALSRR376 pKa = 11.84PGPPGRR382 pKa = 11.84ARR384 pKa = 11.84PGPPDD389 pKa = 3.2QAALTRR395 pKa = 11.84TRR397 pKa = 11.84QTRR400 pKa = 11.84PRR402 pKa = 11.84ATGQAPGGPPDD413 pKa = 4.13PARR416 pKa = 11.84QGARR420 pKa = 11.84QATLARR426 pKa = 11.84TRR428 pKa = 11.84QTRR431 pKa = 11.84PRR433 pKa = 11.84SAAQAAPGPPDD444 pKa = 3.64PARR447 pKa = 11.84QARR450 pKa = 11.84PAALARR456 pKa = 11.84ARR458 pKa = 11.84QPRR461 pKa = 11.84PRR463 pKa = 11.84AASQAAPGPADD474 pKa = 3.79PARR477 pKa = 11.84QAALTRR483 pKa = 11.84ARR485 pKa = 11.84PTRR488 pKa = 11.84PRR490 pKa = 11.84SGPARR495 pKa = 11.84PRR497 pKa = 11.84LPPSPSAAEE506 pKa = 4.0PSRR509 pKa = 11.84ARR511 pKa = 11.84NPASLRR517 pKa = 11.84LPSLAIAPPPAASGPSSRR535 pKa = 11.84PPGLLSTSLRR545 pKa = 11.84ACQPGRR551 pKa = 11.84QPSPCC556 pKa = 3.86
MM1 pKa = 7.18GWAALAGPARR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84VLQTARR20 pKa = 11.84RR21 pKa = 11.84ARR23 pKa = 11.84QTTLSRR29 pKa = 11.84PGPPDD34 pKa = 3.06QTALAGPHH42 pKa = 3.99QTRR45 pKa = 11.84PRR47 pKa = 11.84RR48 pKa = 11.84VLQAALSRR56 pKa = 11.84PGPPDD61 pKa = 2.82QAARR65 pKa = 11.84PRR67 pKa = 11.84PVRR70 pKa = 11.84PRR72 pKa = 11.84SPGRR76 pKa = 11.84ARR78 pKa = 11.84QTRR81 pKa = 11.84PRR83 pKa = 11.84SPRR86 pKa = 11.84PARR89 pKa = 11.84PRR91 pKa = 11.84RR92 pKa = 11.84VLQTALSRR100 pKa = 11.84PGPPDD105 pKa = 3.0RR106 pKa = 11.84AGSSRR111 pKa = 11.84PRR113 pKa = 11.84AGPARR118 pKa = 11.84PRR120 pKa = 11.84AGSSRR125 pKa = 11.84PRR127 pKa = 11.84AGPAGPRR134 pKa = 11.84QAHH137 pKa = 3.87QTARR141 pKa = 11.84ARR143 pKa = 11.84SSRR146 pKa = 11.84PRR148 pKa = 11.84SPGRR152 pKa = 11.84VRR154 pKa = 11.84QTRR157 pKa = 11.84PRR159 pKa = 11.84SLGPPDD165 pKa = 3.69QPEE168 pKa = 4.02LAGPARR174 pKa = 11.84SRR176 pKa = 11.84RR177 pKa = 11.84VLRR180 pKa = 11.84TAPGRR185 pKa = 11.84ARR187 pKa = 11.84QVLQTTVSRR196 pKa = 11.84PGPPNQATRR205 pKa = 11.84AGPTRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84SPGRR216 pKa = 11.84VRR218 pKa = 11.84QTRR221 pKa = 11.84PSSPSQAAPGPSDD234 pKa = 3.35PAPRR238 pKa = 11.84ARR240 pKa = 11.84QAALTRR246 pKa = 11.84ARR248 pKa = 11.84QAAPGHH254 pKa = 6.84PDD256 pKa = 3.06RR257 pKa = 11.84ALQAGSARR265 pKa = 11.84PGRR268 pKa = 11.84ARR270 pKa = 11.84PGPPDD275 pKa = 3.2QTALARR281 pKa = 11.84PQQTRR286 pKa = 11.84PRR288 pKa = 11.84RR289 pKa = 11.84VLQAALSRR297 pKa = 11.84PGPPDD302 pKa = 3.18QATHH306 pKa = 7.16PGPARR311 pKa = 11.84PARR314 pKa = 11.84PRR316 pKa = 11.84PPGRR320 pKa = 11.84APQTGPRR327 pKa = 11.84SPAPARR333 pKa = 11.84PNRR336 pKa = 11.84TCRR339 pKa = 11.84ARR341 pKa = 11.84QAVAGPPDD349 pKa = 3.9RR350 pKa = 11.84GLQAGATRR358 pKa = 11.84PRR360 pKa = 11.84RR361 pKa = 11.84VRR363 pKa = 11.84QTTRR367 pKa = 11.84WALQTALSRR376 pKa = 11.84PGPPGRR382 pKa = 11.84ARR384 pKa = 11.84PGPPDD389 pKa = 3.2QAALTRR395 pKa = 11.84TRR397 pKa = 11.84QTRR400 pKa = 11.84PRR402 pKa = 11.84ATGQAPGGPPDD413 pKa = 4.13PARR416 pKa = 11.84QGARR420 pKa = 11.84QATLARR426 pKa = 11.84TRR428 pKa = 11.84QTRR431 pKa = 11.84PRR433 pKa = 11.84SAAQAAPGPPDD444 pKa = 3.64PARR447 pKa = 11.84QARR450 pKa = 11.84PAALARR456 pKa = 11.84ARR458 pKa = 11.84QPRR461 pKa = 11.84PRR463 pKa = 11.84AASQAAPGPADD474 pKa = 3.79PARR477 pKa = 11.84QAALTRR483 pKa = 11.84ARR485 pKa = 11.84PTRR488 pKa = 11.84PRR490 pKa = 11.84SGPARR495 pKa = 11.84PRR497 pKa = 11.84LPPSPSAAEE506 pKa = 4.0PSRR509 pKa = 11.84ARR511 pKa = 11.84NPASLRR517 pKa = 11.84LPSLAIAPPPAASGPSSRR535 pKa = 11.84PPGLLSTSLRR545 pKa = 11.84ACQPGRR551 pKa = 11.84QPSPCC556 pKa = 3.86
Molecular weight: 58.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
29353646 |
24 |
34744 |
719.8 |
80.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.125 ± 0.013 | 2.092 ± 0.01 |
4.879 ± 0.008 | 7.353 ± 0.015 |
3.412 ± 0.007 | 6.548 ± 0.016 |
2.58 ± 0.005 | 4.119 ± 0.009 |
5.725 ± 0.014 | 9.863 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.073 ± 0.005 | 3.482 ± 0.009 |
6.492 ± 0.02 | 4.961 ± 0.012 |
5.833 ± 0.011 | 8.577 ± 0.014 |
5.266 ± 0.009 | 5.959 ± 0.009 |
1.158 ± 0.004 | 2.493 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
