
Gammaproteobacteria bacterium LSUCC0112
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; unclassified Gammaproteobacteria
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
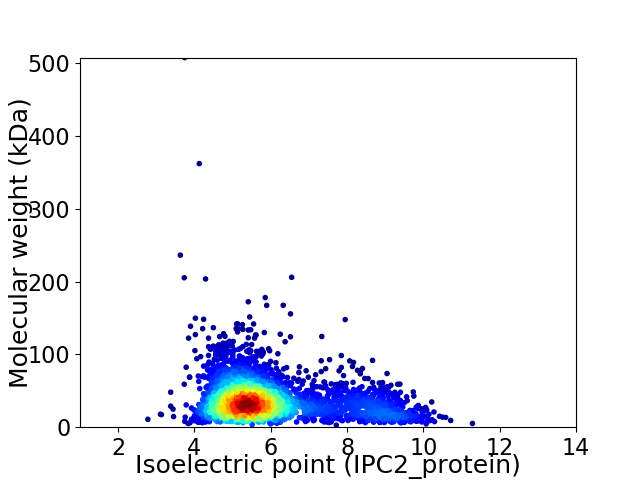
Virtual 2D-PAGE plot for 3053 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8UXX4|A0A4Y8UXX4_9GAMM Chemotaxis protein CheW OS=Gammaproteobacteria bacterium LSUCC0112 OX=1778437 GN=E3V39_08910 PE=4 SV=1
MM1 pKa = 7.6RR2 pKa = 11.84KK3 pKa = 9.24SVPSFNFLTWLYY15 pKa = 10.97GFFVCFSFEE24 pKa = 3.99THH26 pKa = 6.51AQTQNYY32 pKa = 9.67NIDD35 pKa = 3.67DD36 pKa = 3.88TSRR39 pKa = 11.84AAVQVIVDD47 pKa = 3.77NATGSGTLIIVDD59 pKa = 4.12GKK61 pKa = 8.74PTVFTNRR68 pKa = 11.84HH69 pKa = 3.96VVEE72 pKa = 5.25GFDD75 pKa = 3.27QATIAVLVDD84 pKa = 3.88PNAPAEE90 pKa = 4.08PMFIANLVGFSQEE103 pKa = 3.52YY104 pKa = 10.78DD105 pKa = 3.44FAVYY109 pKa = 9.26TLATDD114 pKa = 4.09LQGNTVSAQQLRR126 pKa = 11.84DD127 pKa = 3.03GSFGVRR133 pKa = 11.84FPDD136 pKa = 3.85ITVQDD141 pKa = 3.89IDD143 pKa = 3.97NKK145 pKa = 10.58GSDD148 pKa = 3.57VRR150 pKa = 11.84RR151 pKa = 11.84GDD153 pKa = 3.41TVGIFGYY160 pKa = 9.86PGIGDD165 pKa = 4.25DD166 pKa = 3.83EE167 pKa = 4.54LVYY170 pKa = 8.55TTGIISSVQFGEE182 pKa = 4.4YY183 pKa = 9.57KK184 pKa = 10.68GEE186 pKa = 4.11RR187 pKa = 11.84LPMWYY192 pKa = 8.04RR193 pKa = 11.84TNAEE197 pKa = 4.12MSPGNSGGMALNGRR211 pKa = 11.84GEE213 pKa = 4.65FIGIPTSVRR222 pKa = 11.84TEE224 pKa = 3.76NLTGGRR230 pKa = 11.84LGSLLAVPFVMAILEE245 pKa = 4.38DD246 pKa = 4.23DD247 pKa = 4.09EE248 pKa = 5.75GLVASWSGVDD258 pKa = 3.43SSIAASEE265 pKa = 4.13LNILQEE271 pKa = 4.28PGFGSVTLAPSEE283 pKa = 3.96LGEE286 pKa = 4.13PFFAEE291 pKa = 5.08LISGGSVDD299 pKa = 4.96VSYY302 pKa = 11.2LGNNCVGFAASNPDD316 pKa = 3.26YY317 pKa = 10.8RR318 pKa = 11.84IALTDD323 pKa = 4.13DD324 pKa = 3.64MTDD327 pKa = 3.19MTILFLAEE335 pKa = 4.76DD336 pKa = 4.0EE337 pKa = 4.91GADD340 pKa = 3.23TTLIVNAPDD349 pKa = 4.62GRR351 pKa = 11.84WHH353 pKa = 6.97CNDD356 pKa = 4.85DD357 pKa = 3.69EE358 pKa = 6.58SDD360 pKa = 3.64DD361 pKa = 4.07SRR363 pKa = 11.84DD364 pKa = 3.57PGLLFTNATSGQYY377 pKa = 10.27DD378 pKa = 3.28IWIGSYY384 pKa = 10.12DD385 pKa = 3.56QNDD388 pKa = 3.75AVSGVLAVVDD398 pKa = 5.32ASDD401 pKa = 3.6FDD403 pKa = 4.76NIEE406 pKa = 4.21SSVGAQGTALDD417 pKa = 4.65FAADD421 pKa = 4.43PYY423 pKa = 11.23YY424 pKa = 11.27GSADD428 pKa = 3.41LAAGFTPDD436 pKa = 3.29PHH438 pKa = 7.36VVSVSAGGAVDD449 pKa = 3.92VSIAGYY455 pKa = 10.71GNEE458 pKa = 3.95CRR460 pKa = 11.84GFASSAPDD468 pKa = 3.41YY469 pKa = 10.89KK470 pKa = 10.48VTLSGDD476 pKa = 3.53SALLQLYY483 pKa = 8.33FVASSIGDD491 pKa = 3.79DD492 pKa = 3.11ATLIINNPEE501 pKa = 4.08GTWLCNDD508 pKa = 4.62DD509 pKa = 5.26APSSLNPGITINNPVAGRR527 pKa = 11.84YY528 pKa = 8.99DD529 pKa = 3.16IWVGAYY535 pKa = 9.8AQGEE539 pKa = 4.57FINGEE544 pKa = 3.88LRR546 pKa = 11.84ITEE549 pKa = 4.6LSTTVPP555 pKa = 3.55
MM1 pKa = 7.6RR2 pKa = 11.84KK3 pKa = 9.24SVPSFNFLTWLYY15 pKa = 10.97GFFVCFSFEE24 pKa = 3.99THH26 pKa = 6.51AQTQNYY32 pKa = 9.67NIDD35 pKa = 3.67DD36 pKa = 3.88TSRR39 pKa = 11.84AAVQVIVDD47 pKa = 3.77NATGSGTLIIVDD59 pKa = 4.12GKK61 pKa = 8.74PTVFTNRR68 pKa = 11.84HH69 pKa = 3.96VVEE72 pKa = 5.25GFDD75 pKa = 3.27QATIAVLVDD84 pKa = 3.88PNAPAEE90 pKa = 4.08PMFIANLVGFSQEE103 pKa = 3.52YY104 pKa = 10.78DD105 pKa = 3.44FAVYY109 pKa = 9.26TLATDD114 pKa = 4.09LQGNTVSAQQLRR126 pKa = 11.84DD127 pKa = 3.03GSFGVRR133 pKa = 11.84FPDD136 pKa = 3.85ITVQDD141 pKa = 3.89IDD143 pKa = 3.97NKK145 pKa = 10.58GSDD148 pKa = 3.57VRR150 pKa = 11.84RR151 pKa = 11.84GDD153 pKa = 3.41TVGIFGYY160 pKa = 9.86PGIGDD165 pKa = 4.25DD166 pKa = 3.83EE167 pKa = 4.54LVYY170 pKa = 8.55TTGIISSVQFGEE182 pKa = 4.4YY183 pKa = 9.57KK184 pKa = 10.68GEE186 pKa = 4.11RR187 pKa = 11.84LPMWYY192 pKa = 8.04RR193 pKa = 11.84TNAEE197 pKa = 4.12MSPGNSGGMALNGRR211 pKa = 11.84GEE213 pKa = 4.65FIGIPTSVRR222 pKa = 11.84TEE224 pKa = 3.76NLTGGRR230 pKa = 11.84LGSLLAVPFVMAILEE245 pKa = 4.38DD246 pKa = 4.23DD247 pKa = 4.09EE248 pKa = 5.75GLVASWSGVDD258 pKa = 3.43SSIAASEE265 pKa = 4.13LNILQEE271 pKa = 4.28PGFGSVTLAPSEE283 pKa = 3.96LGEE286 pKa = 4.13PFFAEE291 pKa = 5.08LISGGSVDD299 pKa = 4.96VSYY302 pKa = 11.2LGNNCVGFAASNPDD316 pKa = 3.26YY317 pKa = 10.8RR318 pKa = 11.84IALTDD323 pKa = 4.13DD324 pKa = 3.64MTDD327 pKa = 3.19MTILFLAEE335 pKa = 4.76DD336 pKa = 4.0EE337 pKa = 4.91GADD340 pKa = 3.23TTLIVNAPDD349 pKa = 4.62GRR351 pKa = 11.84WHH353 pKa = 6.97CNDD356 pKa = 4.85DD357 pKa = 3.69EE358 pKa = 6.58SDD360 pKa = 3.64DD361 pKa = 4.07SRR363 pKa = 11.84DD364 pKa = 3.57PGLLFTNATSGQYY377 pKa = 10.27DD378 pKa = 3.28IWIGSYY384 pKa = 10.12DD385 pKa = 3.56QNDD388 pKa = 3.75AVSGVLAVVDD398 pKa = 5.32ASDD401 pKa = 3.6FDD403 pKa = 4.76NIEE406 pKa = 4.21SSVGAQGTALDD417 pKa = 4.65FAADD421 pKa = 4.43PYY423 pKa = 11.23YY424 pKa = 11.27GSADD428 pKa = 3.41LAAGFTPDD436 pKa = 3.29PHH438 pKa = 7.36VVSVSAGGAVDD449 pKa = 3.92VSIAGYY455 pKa = 10.71GNEE458 pKa = 3.95CRR460 pKa = 11.84GFASSAPDD468 pKa = 3.41YY469 pKa = 10.89KK470 pKa = 10.48VTLSGDD476 pKa = 3.53SALLQLYY483 pKa = 8.33FVASSIGDD491 pKa = 3.79DD492 pKa = 3.11ATLIINNPEE501 pKa = 4.08GTWLCNDD508 pKa = 4.62DD509 pKa = 5.26APSSLNPGITINNPVAGRR527 pKa = 11.84YY528 pKa = 8.99DD529 pKa = 3.16IWVGAYY535 pKa = 9.8AQGEE539 pKa = 4.57FINGEE544 pKa = 3.88LRR546 pKa = 11.84ITEE549 pKa = 4.6LSTTVPP555 pKa = 3.55
Molecular weight: 58.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8V434|A0A4Y8V434_9GAMM DsbC domain-containing protein OS=Gammaproteobacteria bacterium LSUCC0112 OX=1778437 GN=E3V39_07115 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.58RR3 pKa = 11.84TFQPSLIKK11 pKa = 10.3RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.96GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84LVIKK32 pKa = 10.33RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84AKK41 pKa = 9.71LTAA44 pKa = 4.28
MM1 pKa = 7.45KK2 pKa = 9.58RR3 pKa = 11.84TFQPSLIKK11 pKa = 10.3RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.96GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84LVIKK32 pKa = 10.33RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84AKK41 pKa = 9.71LTAA44 pKa = 4.28
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1073988 |
23 |
4975 |
351.8 |
38.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.015 ± 0.05 | 0.94 ± 0.015 |
5.738 ± 0.033 | 5.427 ± 0.037 |
3.907 ± 0.032 | 7.488 ± 0.048 |
2.19 ± 0.026 | 5.692 ± 0.034 |
3.231 ± 0.036 | 10.63 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.649 ± 0.023 | 3.773 ± 0.038 |
4.533 ± 0.035 | 4.516 ± 0.043 |
5.951 ± 0.05 | 6.527 ± 0.037 |
5.572 ± 0.071 | 7.238 ± 0.034 |
1.324 ± 0.017 | 2.654 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
