
Phytophthora fragariae
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Phytophthora
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
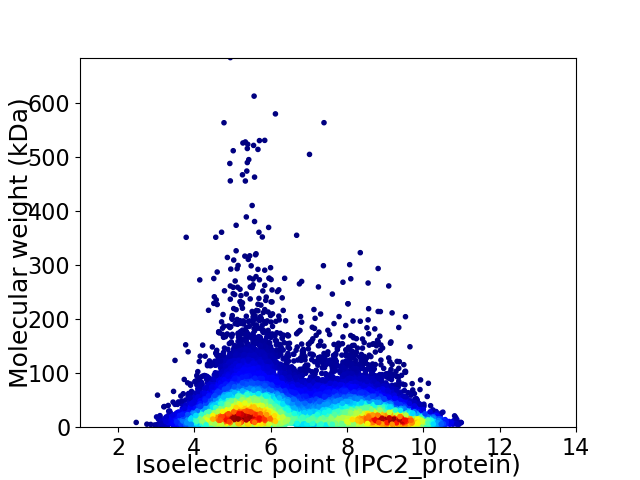
Virtual 2D-PAGE plot for 34421 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A3WLA1|A0A6A3WLA1_9STRA Uncharacterized protein OS=Phytophthora fragariae OX=53985 GN=PF002_g27136 PE=4 SV=1
MM1 pKa = 7.16VNVLRR6 pKa = 11.84TTSALAMAATTFGLANAGSFEE27 pKa = 4.39CTGINYY33 pKa = 9.54NIRR36 pKa = 11.84AGPDD40 pKa = 2.57WATADD45 pKa = 4.08VKK47 pKa = 10.81CKK49 pKa = 9.94PSSQIATEE57 pKa = 4.34LSTLKK62 pKa = 10.88GVTDD66 pKa = 4.4TIRR69 pKa = 11.84LYY71 pKa = 11.34SLTDD75 pKa = 3.55CDD77 pKa = 3.0QATAVVPAAIEE88 pKa = 4.13AGLKK92 pKa = 9.83IEE94 pKa = 4.43LGMWVDD100 pKa = 3.67SATSSFEE107 pKa = 3.84AEE109 pKa = 3.71KK110 pKa = 10.82AAFQTLLEE118 pKa = 4.34TGIVTADD125 pKa = 3.8NIVGIHH131 pKa = 5.63VGSEE135 pKa = 3.25AVYY138 pKa = 10.56RR139 pKa = 11.84GDD141 pKa = 2.94VMAAVAISNLEE152 pKa = 4.18EE153 pKa = 4.55IRR155 pKa = 11.84TLCQANSGAASIPLTIADD173 pKa = 3.89IGDD176 pKa = 3.75TYY178 pKa = 11.24SAYY181 pKa = 9.91PEE183 pKa = 4.32MIQAVDD189 pKa = 3.78YY190 pKa = 11.34VSANYY195 pKa = 9.71FPFWEE200 pKa = 4.59KK201 pKa = 11.15VDD203 pKa = 3.35IDD205 pKa = 3.6GAADD209 pKa = 3.25HH210 pKa = 6.59FYY212 pKa = 11.13EE213 pKa = 4.57RR214 pKa = 11.84FSALVEE220 pKa = 4.16TASTYY225 pKa = 10.07SKK227 pKa = 10.62EE228 pKa = 4.31VIVGEE233 pKa = 4.43TGWASNGTDD242 pKa = 3.57ADD244 pKa = 3.89ASPATAANAAKK255 pKa = 10.04YY256 pKa = 9.66FHH258 pKa = 7.44DD259 pKa = 5.13FYY261 pKa = 11.39TLAEE265 pKa = 4.66EE266 pKa = 4.29KK267 pKa = 10.79DD268 pKa = 3.24LMYY271 pKa = 10.32FYY273 pKa = 10.58FSAFDD278 pKa = 4.17EE279 pKa = 4.52PWKK282 pKa = 10.71LATLEE287 pKa = 4.08ANEE290 pKa = 4.12TVEE293 pKa = 5.12AYY295 pKa = 10.4FGLFTQEE302 pKa = 4.22GVLKK306 pKa = 10.51DD307 pKa = 3.94VISAATASLDD317 pKa = 3.65GSDD320 pKa = 3.87SATITAVGADD330 pKa = 4.01SSTSDD335 pKa = 3.0ATTTASAASTSADD348 pKa = 3.09NSTLITTTSVYY359 pKa = 9.66ATSSDD364 pKa = 4.2DD365 pKa = 3.69DD366 pKa = 5.13DD367 pKa = 6.08SIQGEE372 pKa = 4.24ATSDD376 pKa = 3.11VDD378 pKa = 3.68ASSDD382 pKa = 3.76EE383 pKa = 4.29EE384 pKa = 4.52TPSDD388 pKa = 4.89DD389 pKa = 5.62GEE391 pKa = 6.28DD392 pKa = 3.63DD393 pKa = 3.9TTDD396 pKa = 3.17STTQTSTTTTSSSHH410 pKa = 6.64KK411 pKa = 10.27DD412 pKa = 3.09CGMM415 pKa = 3.95
MM1 pKa = 7.16VNVLRR6 pKa = 11.84TTSALAMAATTFGLANAGSFEE27 pKa = 4.39CTGINYY33 pKa = 9.54NIRR36 pKa = 11.84AGPDD40 pKa = 2.57WATADD45 pKa = 4.08VKK47 pKa = 10.81CKK49 pKa = 9.94PSSQIATEE57 pKa = 4.34LSTLKK62 pKa = 10.88GVTDD66 pKa = 4.4TIRR69 pKa = 11.84LYY71 pKa = 11.34SLTDD75 pKa = 3.55CDD77 pKa = 3.0QATAVVPAAIEE88 pKa = 4.13AGLKK92 pKa = 9.83IEE94 pKa = 4.43LGMWVDD100 pKa = 3.67SATSSFEE107 pKa = 3.84AEE109 pKa = 3.71KK110 pKa = 10.82AAFQTLLEE118 pKa = 4.34TGIVTADD125 pKa = 3.8NIVGIHH131 pKa = 5.63VGSEE135 pKa = 3.25AVYY138 pKa = 10.56RR139 pKa = 11.84GDD141 pKa = 2.94VMAAVAISNLEE152 pKa = 4.18EE153 pKa = 4.55IRR155 pKa = 11.84TLCQANSGAASIPLTIADD173 pKa = 3.89IGDD176 pKa = 3.75TYY178 pKa = 11.24SAYY181 pKa = 9.91PEE183 pKa = 4.32MIQAVDD189 pKa = 3.78YY190 pKa = 11.34VSANYY195 pKa = 9.71FPFWEE200 pKa = 4.59KK201 pKa = 11.15VDD203 pKa = 3.35IDD205 pKa = 3.6GAADD209 pKa = 3.25HH210 pKa = 6.59FYY212 pKa = 11.13EE213 pKa = 4.57RR214 pKa = 11.84FSALVEE220 pKa = 4.16TASTYY225 pKa = 10.07SKK227 pKa = 10.62EE228 pKa = 4.31VIVGEE233 pKa = 4.43TGWASNGTDD242 pKa = 3.57ADD244 pKa = 3.89ASPATAANAAKK255 pKa = 10.04YY256 pKa = 9.66FHH258 pKa = 7.44DD259 pKa = 5.13FYY261 pKa = 11.39TLAEE265 pKa = 4.66EE266 pKa = 4.29KK267 pKa = 10.79DD268 pKa = 3.24LMYY271 pKa = 10.32FYY273 pKa = 10.58FSAFDD278 pKa = 4.17EE279 pKa = 4.52PWKK282 pKa = 10.71LATLEE287 pKa = 4.08ANEE290 pKa = 4.12TVEE293 pKa = 5.12AYY295 pKa = 10.4FGLFTQEE302 pKa = 4.22GVLKK306 pKa = 10.51DD307 pKa = 3.94VISAATASLDD317 pKa = 3.65GSDD320 pKa = 3.87SATITAVGADD330 pKa = 4.01SSTSDD335 pKa = 3.0ATTTASAASTSADD348 pKa = 3.09NSTLITTTSVYY359 pKa = 9.66ATSSDD364 pKa = 4.2DD365 pKa = 3.69DD366 pKa = 5.13DD367 pKa = 6.08SIQGEE372 pKa = 4.24ATSDD376 pKa = 3.11VDD378 pKa = 3.68ASSDD382 pKa = 3.76EE383 pKa = 4.29EE384 pKa = 4.52TPSDD388 pKa = 4.89DD389 pKa = 5.62GEE391 pKa = 6.28DD392 pKa = 3.63DD393 pKa = 3.9TTDD396 pKa = 3.17STTQTSTTTTSSSHH410 pKa = 6.64KK411 pKa = 10.27DD412 pKa = 3.09CGMM415 pKa = 3.95
Molecular weight: 43.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A3YZK2|A0A6A3YZK2_9STRA Uncharacterized protein OS=Phytophthora fragariae OX=53985 GN=PF001_g8246 PE=4 SV=1
MM1 pKa = 7.51LRR3 pKa = 11.84GCRR6 pKa = 11.84RR7 pKa = 11.84SLRR10 pKa = 11.84LLRR13 pKa = 11.84VIRR16 pKa = 11.84MLLGSRR22 pKa = 11.84RR23 pKa = 11.84VLRR26 pKa = 11.84SLWIARR32 pKa = 11.84VIRR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84LSLLFAWSRR50 pKa = 11.84RR51 pKa = 11.84VGVMLGRR58 pKa = 11.84ASGPRR63 pKa = 11.84ALGAARR69 pKa = 11.84TMIWLSVVMLGGPP82 pKa = 3.5
MM1 pKa = 7.51LRR3 pKa = 11.84GCRR6 pKa = 11.84RR7 pKa = 11.84SLRR10 pKa = 11.84LLRR13 pKa = 11.84VIRR16 pKa = 11.84MLLGSRR22 pKa = 11.84RR23 pKa = 11.84VLRR26 pKa = 11.84SLWIARR32 pKa = 11.84VIRR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84LSLLFAWSRR50 pKa = 11.84RR51 pKa = 11.84VGVMLGRR58 pKa = 11.84ASGPRR63 pKa = 11.84ALGAARR69 pKa = 11.84TMIWLSVVMLGGPP82 pKa = 3.5
Molecular weight: 9.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11269359 |
12 |
10552 |
327.4 |
36.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.578 ± 0.016 | 1.656 ± 0.008 |
5.77 ± 0.01 | 6.622 ± 0.019 |
3.512 ± 0.01 | 6.155 ± 0.015 |
2.307 ± 0.007 | 3.709 ± 0.009 |
4.919 ± 0.016 | 9.179 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.426 ± 0.007 | 3.25 ± 0.007 |
5.044 ± 0.012 | 4.155 ± 0.011 |
6.691 ± 0.017 | 8.236 ± 0.019 |
5.912 ± 0.015 | 7.182 ± 0.014 |
1.244 ± 0.005 | 2.451 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
