
Haloredivivus sp. (strain G17)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Nanohaloarchaea; Candidatus Haloredivivus
Average proteome isoelectric point is 5.44
Get precalculated fractions of proteins
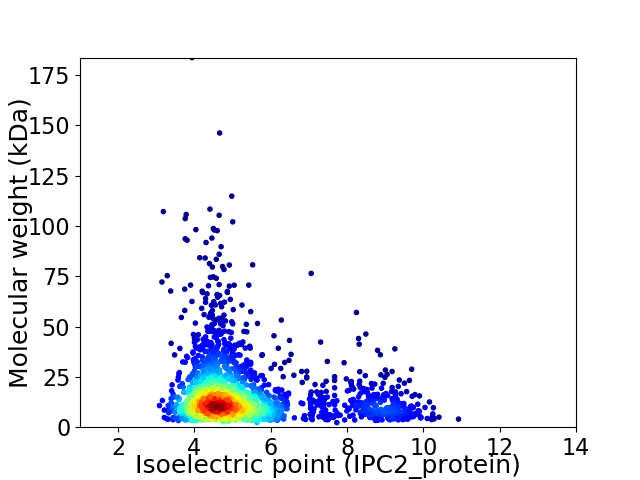
Virtual 2D-PAGE plot for 2152 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|H0AAF9|H0AAF9_HALSG Uncharacterized protein OS=Haloredivivus sp. (strain G17) OX=1072681 GN=HRED_05089 PE=4 SV=1
MM1 pKa = 7.57IKK3 pKa = 10.45LLRR6 pKa = 11.84LRR8 pKa = 11.84RR9 pKa = 11.84PIVGEE14 pKa = 3.78IAIKK18 pKa = 10.24RR19 pKa = 11.84YY20 pKa = 9.37FLTLLSILLFISPVMAQTYY39 pKa = 10.38DD40 pKa = 3.55FNNCGSLGKK49 pKa = 9.92NGPSSCSYY57 pKa = 10.24TGDD60 pKa = 3.11NSVVINNQGFQKK72 pKa = 8.53WTVPEE77 pKa = 4.05SGKK80 pKa = 10.33YY81 pKa = 9.39RR82 pKa = 11.84IKK84 pKa = 10.91AAGAQGAAGDD94 pKa = 3.59PSYY97 pKa = 10.99RR98 pKa = 11.84GGYY101 pKa = 8.22GALVKK106 pKa = 10.87GSFNLRR112 pKa = 11.84EE113 pKa = 4.3GEE115 pKa = 4.43DD116 pKa = 3.13IVIAVGQKK124 pKa = 8.44GTAGSPSTNDD134 pKa = 2.94NDD136 pKa = 4.12GGGGGGSFVVSSSDD150 pKa = 3.41YY151 pKa = 10.11TSASTSDD158 pKa = 3.11ILVIAGGGAGTRR170 pKa = 11.84AYY172 pKa = 11.12VNDD175 pKa = 3.69NGCPGNEE182 pKa = 3.62GEE184 pKa = 4.38YY185 pKa = 10.29AIEE188 pKa = 4.8GSGSDD193 pKa = 3.5QTSSCPVKK201 pKa = 10.5STDD204 pKa = 3.52LGEE207 pKa = 4.34GGIVSSGSWGSAGGGWGSNGEE228 pKa = 3.99GDD230 pKa = 4.08YY231 pKa = 10.07DD232 pKa = 3.76TSAGGRR238 pKa = 11.84SWFNGLEE245 pKa = 4.23GGHH248 pKa = 6.73DD249 pKa = 4.02SEE251 pKa = 5.76SPPAPGGFGAGGSGHH266 pKa = 6.35GSWGGGGGGGYY277 pKa = 10.02SGGDD281 pKa = 3.51GGRR284 pKa = 11.84VAGGGGSYY292 pKa = 8.84NTGSDD297 pKa = 3.24QSNTGGEE304 pKa = 4.29NSGHH308 pKa = 6.24GYY310 pKa = 10.2VEE312 pKa = 4.16IEE314 pKa = 4.39TVAVTSDD321 pKa = 3.47PTVQSPTYY329 pKa = 8.94TDD331 pKa = 3.32SQATSSSPASFEE343 pKa = 3.26VDD345 pKa = 2.96APVDD349 pKa = 3.14VDD351 pKa = 3.64TSVDD355 pKa = 3.46GGNSLSSCSVDD366 pKa = 3.39VTGDD370 pKa = 3.77DD371 pKa = 3.61NGGSYY376 pKa = 10.65SPSTTVSGDD385 pKa = 3.11TCEE388 pKa = 4.92FSVDD392 pKa = 4.34NNDD395 pKa = 3.8DD396 pKa = 4.37SNWEE400 pKa = 3.91PGEE403 pKa = 4.22TVSVDD408 pKa = 2.86ITATDD413 pKa = 3.66DD414 pKa = 4.53LGNTDD419 pKa = 2.93VGYY422 pKa = 7.86FTHH425 pKa = 7.01TFDD428 pKa = 4.49NTPPTINSVTPSDD441 pKa = 3.87GASSVWGGEE450 pKa = 3.96NGVSVQADD458 pKa = 3.58VVDD461 pKa = 4.83FEE463 pKa = 5.25SVHH466 pKa = 6.18TNDD469 pKa = 4.74DD470 pKa = 3.68LTVDD474 pKa = 4.83FVDD477 pKa = 5.4DD478 pKa = 3.84SLGQFGSVTVSNGDD492 pKa = 3.07GTASVNWEE500 pKa = 3.9RR501 pKa = 11.84NGQQDD506 pKa = 3.03MFGFDD511 pKa = 3.29RR512 pKa = 11.84GTSYY516 pKa = 10.74GWKK519 pKa = 10.28VNVDD523 pKa = 3.78DD524 pKa = 4.41VTASTTTQSQSFTVLSEE541 pKa = 4.09ASTSNPSPTGGDD553 pKa = 3.06SADD556 pKa = 3.58QMKK559 pKa = 10.56VDD561 pKa = 3.83IDD563 pKa = 3.86NPAVSNDD570 pKa = 2.97GSLSGSTVEE579 pKa = 4.49FWVDD583 pKa = 3.37DD584 pKa = 3.93GLEE587 pKa = 4.2VYY589 pKa = 9.64QVNTDD594 pKa = 3.62TVSGSSGTASISLQDD609 pKa = 3.35FEE611 pKa = 5.5FRR613 pKa = 11.84NEE615 pKa = 4.04PADD618 pKa = 3.69GTGDD622 pKa = 3.19WFVKK626 pKa = 10.78VNDD629 pKa = 4.37TYY631 pKa = 11.79SEE633 pKa = 4.05TDD635 pKa = 3.39YY636 pKa = 11.59SRR638 pKa = 11.84SKK640 pKa = 10.95YY641 pKa = 10.67GSNTYY646 pKa = 10.38NFSAAKK652 pKa = 9.48PPLILSEE659 pKa = 5.02DD660 pKa = 3.78PSSYY664 pKa = 10.45PIAEE668 pKa = 4.31TGVGG672 pKa = 3.41
MM1 pKa = 7.57IKK3 pKa = 10.45LLRR6 pKa = 11.84LRR8 pKa = 11.84RR9 pKa = 11.84PIVGEE14 pKa = 3.78IAIKK18 pKa = 10.24RR19 pKa = 11.84YY20 pKa = 9.37FLTLLSILLFISPVMAQTYY39 pKa = 10.38DD40 pKa = 3.55FNNCGSLGKK49 pKa = 9.92NGPSSCSYY57 pKa = 10.24TGDD60 pKa = 3.11NSVVINNQGFQKK72 pKa = 8.53WTVPEE77 pKa = 4.05SGKK80 pKa = 10.33YY81 pKa = 9.39RR82 pKa = 11.84IKK84 pKa = 10.91AAGAQGAAGDD94 pKa = 3.59PSYY97 pKa = 10.99RR98 pKa = 11.84GGYY101 pKa = 8.22GALVKK106 pKa = 10.87GSFNLRR112 pKa = 11.84EE113 pKa = 4.3GEE115 pKa = 4.43DD116 pKa = 3.13IVIAVGQKK124 pKa = 8.44GTAGSPSTNDD134 pKa = 2.94NDD136 pKa = 4.12GGGGGGSFVVSSSDD150 pKa = 3.41YY151 pKa = 10.11TSASTSDD158 pKa = 3.11ILVIAGGGAGTRR170 pKa = 11.84AYY172 pKa = 11.12VNDD175 pKa = 3.69NGCPGNEE182 pKa = 3.62GEE184 pKa = 4.38YY185 pKa = 10.29AIEE188 pKa = 4.8GSGSDD193 pKa = 3.5QTSSCPVKK201 pKa = 10.5STDD204 pKa = 3.52LGEE207 pKa = 4.34GGIVSSGSWGSAGGGWGSNGEE228 pKa = 3.99GDD230 pKa = 4.08YY231 pKa = 10.07DD232 pKa = 3.76TSAGGRR238 pKa = 11.84SWFNGLEE245 pKa = 4.23GGHH248 pKa = 6.73DD249 pKa = 4.02SEE251 pKa = 5.76SPPAPGGFGAGGSGHH266 pKa = 6.35GSWGGGGGGGYY277 pKa = 10.02SGGDD281 pKa = 3.51GGRR284 pKa = 11.84VAGGGGSYY292 pKa = 8.84NTGSDD297 pKa = 3.24QSNTGGEE304 pKa = 4.29NSGHH308 pKa = 6.24GYY310 pKa = 10.2VEE312 pKa = 4.16IEE314 pKa = 4.39TVAVTSDD321 pKa = 3.47PTVQSPTYY329 pKa = 8.94TDD331 pKa = 3.32SQATSSSPASFEE343 pKa = 3.26VDD345 pKa = 2.96APVDD349 pKa = 3.14VDD351 pKa = 3.64TSVDD355 pKa = 3.46GGNSLSSCSVDD366 pKa = 3.39VTGDD370 pKa = 3.77DD371 pKa = 3.61NGGSYY376 pKa = 10.65SPSTTVSGDD385 pKa = 3.11TCEE388 pKa = 4.92FSVDD392 pKa = 4.34NNDD395 pKa = 3.8DD396 pKa = 4.37SNWEE400 pKa = 3.91PGEE403 pKa = 4.22TVSVDD408 pKa = 2.86ITATDD413 pKa = 3.66DD414 pKa = 4.53LGNTDD419 pKa = 2.93VGYY422 pKa = 7.86FTHH425 pKa = 7.01TFDD428 pKa = 4.49NTPPTINSVTPSDD441 pKa = 3.87GASSVWGGEE450 pKa = 3.96NGVSVQADD458 pKa = 3.58VVDD461 pKa = 4.83FEE463 pKa = 5.25SVHH466 pKa = 6.18TNDD469 pKa = 4.74DD470 pKa = 3.68LTVDD474 pKa = 4.83FVDD477 pKa = 5.4DD478 pKa = 3.84SLGQFGSVTVSNGDD492 pKa = 3.07GTASVNWEE500 pKa = 3.9RR501 pKa = 11.84NGQQDD506 pKa = 3.03MFGFDD511 pKa = 3.29RR512 pKa = 11.84GTSYY516 pKa = 10.74GWKK519 pKa = 10.28VNVDD523 pKa = 3.78DD524 pKa = 4.41VTASTTTQSQSFTVLSEE541 pKa = 4.09ASTSNPSPTGGDD553 pKa = 3.06SADD556 pKa = 3.58QMKK559 pKa = 10.56VDD561 pKa = 3.83IDD563 pKa = 3.86NPAVSNDD570 pKa = 2.97GSLSGSTVEE579 pKa = 4.49FWVDD583 pKa = 3.37DD584 pKa = 3.93GLEE587 pKa = 4.2VYY589 pKa = 9.64QVNTDD594 pKa = 3.62TVSGSSGTASISLQDD609 pKa = 3.35FEE611 pKa = 5.5FRR613 pKa = 11.84NEE615 pKa = 4.04PADD618 pKa = 3.69GTGDD622 pKa = 3.19WFVKK626 pKa = 10.78VNDD629 pKa = 4.37TYY631 pKa = 11.79SEE633 pKa = 4.05TDD635 pKa = 3.39YY636 pKa = 11.59SRR638 pKa = 11.84SKK640 pKa = 10.95YY641 pKa = 10.67GSNTYY646 pKa = 10.38NFSAAKK652 pKa = 9.48PPLILSEE659 pKa = 5.02DD660 pKa = 3.78PSSYY664 pKa = 10.45PIAEE668 pKa = 4.31TGVGG672 pKa = 3.41
Molecular weight: 68.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H0ADY3|H0ADY3_HALSG Metalloenzyme domain-containing protein OS=Haloredivivus sp. (strain G17) OX=1072681 GN=HRED_10157 PE=4 SV=1
MM1 pKa = 7.58RR2 pKa = 11.84KK3 pKa = 9.07VRR5 pKa = 11.84RR6 pKa = 11.84LKK8 pKa = 9.76KK9 pKa = 9.11TSLKK13 pKa = 10.6RR14 pKa = 11.84NLGKK18 pKa = 9.98MSQNKK23 pKa = 8.77MNLKK27 pKa = 8.93VKK29 pKa = 10.02AWRR32 pKa = 11.84VV33 pKa = 3.23
MM1 pKa = 7.58RR2 pKa = 11.84KK3 pKa = 9.07VRR5 pKa = 11.84RR6 pKa = 11.84LKK8 pKa = 9.76KK9 pKa = 9.11TSLKK13 pKa = 10.6RR14 pKa = 11.84NLGKK18 pKa = 9.98MSQNKK23 pKa = 8.77MNLKK27 pKa = 8.93VKK29 pKa = 10.02AWRR32 pKa = 11.84VV33 pKa = 3.23
Molecular weight: 4.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
324344 |
20 |
1684 |
150.7 |
17.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.939 ± 0.059 | 0.656 ± 0.021 |
6.901 ± 0.073 | 11.257 ± 0.115 |
4.104 ± 0.045 | 6.864 ± 0.069 |
1.59 ± 0.029 | 6.617 ± 0.06 |
6.176 ± 0.084 | 8.595 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.591 ± 0.036 | 4.423 ± 0.056 |
3.304 ± 0.041 | 3.343 ± 0.043 |
4.869 ± 0.054 | 7.133 ± 0.068 |
4.856 ± 0.055 | 6.599 ± 0.061 |
0.864 ± 0.018 | 3.24 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
