
Mycoplasma suis (strain Illinois)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma; Mycoplasma suis
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
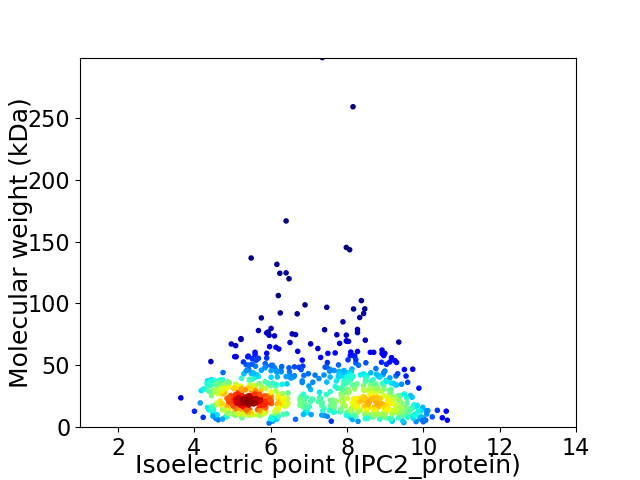
Virtual 2D-PAGE plot for 844 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
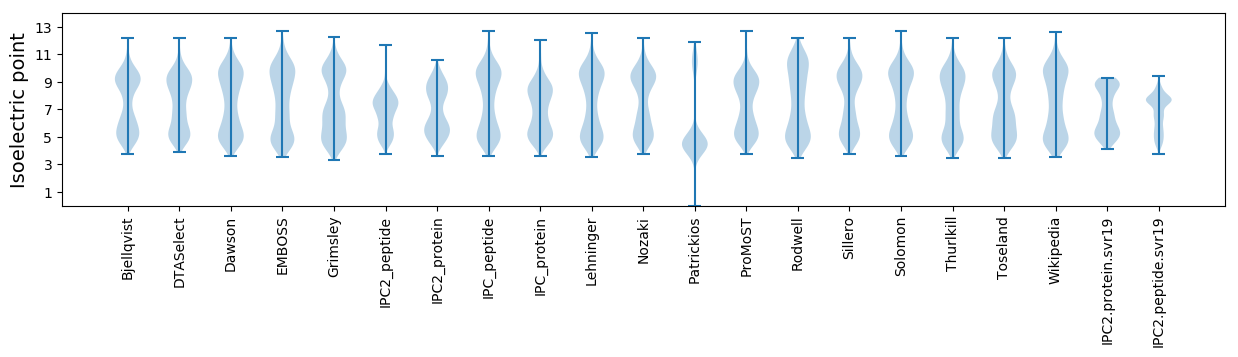
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0QS33|F0QS33_MYCSL Uncharacterized protein OS=Mycoplasma suis (strain Illinois) OX=768700 GN=MSU_0778 PE=4 SV=1
MM1 pKa = 7.39EE2 pKa = 6.16AGFAGDD8 pKa = 3.25WTGCEE13 pKa = 4.2ILWLTASLLTLWEE26 pKa = 4.67SEE28 pKa = 4.39FLWVSSVFVNFSSFLTSLTTCGLLFFEE55 pKa = 4.59SWLWTDD61 pKa = 3.27EE62 pKa = 4.3AGEE65 pKa = 4.24SFAWTEE71 pKa = 4.48SPCGLSDD78 pKa = 3.75SFLINSLCDD87 pKa = 3.08WLLPSNEE94 pKa = 4.42PKK96 pKa = 10.11PEE98 pKa = 4.1PMPPPRR104 pKa = 11.84NLRR107 pKa = 11.84KK108 pKa = 10.04LIPLILL114 pKa = 4.75
MM1 pKa = 7.39EE2 pKa = 6.16AGFAGDD8 pKa = 3.25WTGCEE13 pKa = 4.2ILWLTASLLTLWEE26 pKa = 4.67SEE28 pKa = 4.39FLWVSSVFVNFSSFLTSLTTCGLLFFEE55 pKa = 4.59SWLWTDD61 pKa = 3.27EE62 pKa = 4.3AGEE65 pKa = 4.24SFAWTEE71 pKa = 4.48SPCGLSDD78 pKa = 3.75SFLINSLCDD87 pKa = 3.08WLLPSNEE94 pKa = 4.42PKK96 pKa = 10.11PEE98 pKa = 4.1PMPPPRR104 pKa = 11.84NLRR107 pKa = 11.84KK108 pKa = 10.04LIPLILL114 pKa = 4.75
Molecular weight: 12.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0QR51|F0QR51_MYCSL 50S ribosomal protein L18 OS=Mycoplasma suis (strain Illinois) OX=768700 GN=rplR PE=4 SV=1
MM1 pKa = 7.85AIKK4 pKa = 10.36QKK6 pKa = 10.2VPRR9 pKa = 11.84SSGRR13 pKa = 11.84RR14 pKa = 11.84VVISIDD20 pKa = 3.46YY21 pKa = 10.88NKK23 pKa = 9.66TLTKK27 pKa = 10.5DD28 pKa = 3.46RR29 pKa = 11.84PHH31 pKa = 6.84KK32 pKa = 10.37PLLTKK37 pKa = 10.54LKK39 pKa = 10.38KK40 pKa = 9.72HH41 pKa = 6.34AGRR44 pKa = 11.84NNTGQITVRR53 pKa = 11.84HH54 pKa = 5.99RR55 pKa = 11.84GGEE58 pKa = 3.95NKK60 pKa = 9.74RR61 pKa = 11.84KK62 pKa = 9.89YY63 pKa = 10.46RR64 pKa = 11.84IIDD67 pKa = 3.8FKK69 pKa = 10.99RR70 pKa = 11.84DD71 pKa = 3.21KK72 pKa = 11.29LNVKK76 pKa = 10.03GVVQTIEE83 pKa = 3.83YY84 pKa = 10.1DD85 pKa = 3.5PNRR88 pKa = 11.84TCFISLVHH96 pKa = 5.98YY97 pKa = 10.95ADD99 pKa = 3.63GEE101 pKa = 4.12KK102 pKa = 10.17RR103 pKa = 11.84YY104 pKa = 9.97ILAPSEE110 pKa = 4.14IKK112 pKa = 10.64KK113 pKa = 10.8GDD115 pKa = 3.41IVVSSMEE122 pKa = 4.06RR123 pKa = 11.84TNLRR127 pKa = 11.84IGDD130 pKa = 4.1CMPIGVIPEE139 pKa = 3.56GMMVHH144 pKa = 6.73NIEE147 pKa = 4.64IKK149 pKa = 10.53PGDD152 pKa = 3.78GGKK155 pKa = 8.64MVRR158 pKa = 11.84SAGSQAQILGKK169 pKa = 10.15DD170 pKa = 3.32LTGKK174 pKa = 8.41YY175 pKa = 8.9TIIKK179 pKa = 9.66LPSKK183 pKa = 9.15EE184 pKa = 3.71VRR186 pKa = 11.84KK187 pKa = 10.26VINGARR193 pKa = 11.84ATIGQLSAIEE203 pKa = 4.31HH204 pKa = 5.65NLVRR208 pKa = 11.84LGTAGKK214 pKa = 9.46NRR216 pKa = 11.84HH217 pKa = 5.69LGIRR221 pKa = 11.84PTVRR225 pKa = 11.84GVAMNPNDD233 pKa = 4.55HH234 pKa = 6.27IHH236 pKa = 6.44GGGEE240 pKa = 4.26GRR242 pKa = 11.84CSIGRR247 pKa = 11.84DD248 pKa = 3.47APRR251 pKa = 11.84TPWGKK256 pKa = 9.41RR257 pKa = 11.84HH258 pKa = 6.37LGVKK262 pKa = 8.64TRR264 pKa = 11.84SKK266 pKa = 10.6KK267 pKa = 9.81RR268 pKa = 11.84YY269 pKa = 7.02SNRR272 pKa = 11.84MIVQRR277 pKa = 11.84RR278 pKa = 11.84NAKK281 pKa = 9.37NN282 pKa = 3.0
MM1 pKa = 7.85AIKK4 pKa = 10.36QKK6 pKa = 10.2VPRR9 pKa = 11.84SSGRR13 pKa = 11.84RR14 pKa = 11.84VVISIDD20 pKa = 3.46YY21 pKa = 10.88NKK23 pKa = 9.66TLTKK27 pKa = 10.5DD28 pKa = 3.46RR29 pKa = 11.84PHH31 pKa = 6.84KK32 pKa = 10.37PLLTKK37 pKa = 10.54LKK39 pKa = 10.38KK40 pKa = 9.72HH41 pKa = 6.34AGRR44 pKa = 11.84NNTGQITVRR53 pKa = 11.84HH54 pKa = 5.99RR55 pKa = 11.84GGEE58 pKa = 3.95NKK60 pKa = 9.74RR61 pKa = 11.84KK62 pKa = 9.89YY63 pKa = 10.46RR64 pKa = 11.84IIDD67 pKa = 3.8FKK69 pKa = 10.99RR70 pKa = 11.84DD71 pKa = 3.21KK72 pKa = 11.29LNVKK76 pKa = 10.03GVVQTIEE83 pKa = 3.83YY84 pKa = 10.1DD85 pKa = 3.5PNRR88 pKa = 11.84TCFISLVHH96 pKa = 5.98YY97 pKa = 10.95ADD99 pKa = 3.63GEE101 pKa = 4.12KK102 pKa = 10.17RR103 pKa = 11.84YY104 pKa = 9.97ILAPSEE110 pKa = 4.14IKK112 pKa = 10.64KK113 pKa = 10.8GDD115 pKa = 3.41IVVSSMEE122 pKa = 4.06RR123 pKa = 11.84TNLRR127 pKa = 11.84IGDD130 pKa = 4.1CMPIGVIPEE139 pKa = 3.56GMMVHH144 pKa = 6.73NIEE147 pKa = 4.64IKK149 pKa = 10.53PGDD152 pKa = 3.78GGKK155 pKa = 8.64MVRR158 pKa = 11.84SAGSQAQILGKK169 pKa = 10.15DD170 pKa = 3.32LTGKK174 pKa = 8.41YY175 pKa = 8.9TIIKK179 pKa = 9.66LPSKK183 pKa = 9.15EE184 pKa = 3.71VRR186 pKa = 11.84KK187 pKa = 10.26VINGARR193 pKa = 11.84ATIGQLSAIEE203 pKa = 4.31HH204 pKa = 5.65NLVRR208 pKa = 11.84LGTAGKK214 pKa = 9.46NRR216 pKa = 11.84HH217 pKa = 5.69LGIRR221 pKa = 11.84PTVRR225 pKa = 11.84GVAMNPNDD233 pKa = 4.55HH234 pKa = 6.27IHH236 pKa = 6.44GGGEE240 pKa = 4.26GRR242 pKa = 11.84CSIGRR247 pKa = 11.84DD248 pKa = 3.47APRR251 pKa = 11.84TPWGKK256 pKa = 9.41RR257 pKa = 11.84HH258 pKa = 6.37LGVKK262 pKa = 8.64TRR264 pKa = 11.84SKK266 pKa = 10.6KK267 pKa = 9.81RR268 pKa = 11.84YY269 pKa = 7.02SNRR272 pKa = 11.84MIVQRR277 pKa = 11.84RR278 pKa = 11.84NAKK281 pKa = 9.37NN282 pKa = 3.0
Molecular weight: 31.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
219786 |
33 |
2643 |
260.4 |
29.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.647 ± 0.074 | 1.316 ± 0.038 |
4.465 ± 0.067 | 9.19 ± 0.101 |
5.234 ± 0.096 | 6.018 ± 0.097 |
1.281 ± 0.033 | 7.075 ± 0.089 |
10.23 ± 0.093 | 9.722 ± 0.102 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.536 ± 0.025 | 6.056 ± 0.067 |
2.767 ± 0.043 | 3.701 ± 0.062 |
3.723 ± 0.054 | 9.659 ± 0.106 |
4.468 ± 0.056 | 5.084 ± 0.068 |
1.703 ± 0.043 | 3.126 ± 0.055 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
