
Flavobacterium sp. 102
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; unclassified Flavobacterium
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
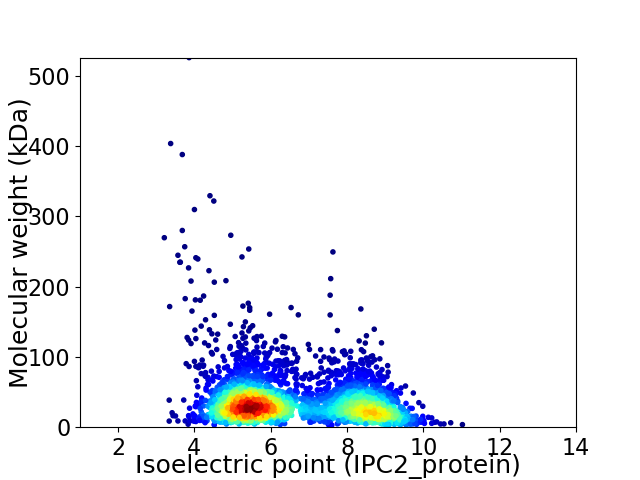
Virtual 2D-PAGE plot for 3318 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A495KNL4|A0A495KNL4_9FLAO Dihydroorotase OS=Flavobacterium sp. 102 OX=2135623 GN=C8C84_2407 PE=4 SV=1
MM1 pKa = 7.55KK2 pKa = 10.52KK3 pKa = 10.2NLFFLLFIAFLTTNCFSQFSKK24 pKa = 8.35THH26 pKa = 6.21YY27 pKa = 9.98IPPLSGSSNVTAEE40 pKa = 4.07DD41 pKa = 3.28QFIYY45 pKa = 10.54ISTPNINPINFRR57 pKa = 11.84IIEE60 pKa = 4.2LGGSVINGSVSRR72 pKa = 11.84STPFIYY78 pKa = 10.64NIGFGNDD85 pKa = 3.19TQLHH89 pKa = 5.47VDD91 pKa = 3.93ASQTNNTFSNKK102 pKa = 9.49GYY104 pKa = 10.18IIEE107 pKa = 4.9ADD109 pKa = 3.49DD110 pKa = 5.02LIYY113 pKa = 11.18VSARR117 pKa = 11.84VTAGNGNHH125 pKa = 6.81AGEE128 pKa = 4.55LVSKK132 pKa = 10.68GKK134 pKa = 10.6ASLGNRR140 pKa = 11.84FRR142 pKa = 11.84IGAITNLTVDD152 pKa = 4.34NYY154 pKa = 11.39NDD156 pKa = 2.8IHH158 pKa = 5.5TTFISVLASEE168 pKa = 5.02NNTTVNFSDD177 pKa = 4.75FKK179 pKa = 11.12PGVQLINSSTGDD191 pKa = 3.53SPFSIVLNSGEE202 pKa = 4.39SYY204 pKa = 10.89VIAVQGPLNPNRR216 pKa = 11.84DD217 pKa = 3.58GLIGSLVTSDD227 pKa = 4.84KK228 pKa = 10.9PIALNCGSFGGSNAAGNLDD247 pKa = 3.99LGFDD251 pKa = 3.68QIVPAEE257 pKa = 4.17RR258 pKa = 11.84INNNEE263 pKa = 3.96YY264 pKa = 10.54IFIKK268 pKa = 9.68STGVDD273 pKa = 2.85IVEE276 pKa = 4.46TVLLVADD283 pKa = 3.93VDD285 pKa = 3.86NTEE288 pKa = 4.12VFLSGSTTPDD298 pKa = 3.07VILNAGNYY306 pKa = 9.92AVFNGSSFDD315 pKa = 4.23LNGNLYY321 pKa = 9.95IRR323 pKa = 11.84TNQKK327 pKa = 9.19VFAYY331 pKa = 7.92QTIGDD336 pKa = 4.23NSASDD341 pKa = 3.44QRR343 pKa = 11.84NQEE346 pKa = 4.12MFFVPPLSCQTPRR359 pKa = 11.84IIDD362 pKa = 4.21NIPSIDD368 pKa = 3.8YY369 pKa = 10.28VGSRR373 pKa = 11.84QFTGRR378 pKa = 11.84VTIVTKK384 pKa = 10.37TGASLNFIIDD394 pKa = 3.59GTNYY398 pKa = 9.56TLAEE402 pKa = 4.32LSLITNITGPTNVVGNNNYY421 pKa = 6.19QTYY424 pKa = 10.57VITGLSGDD432 pKa = 3.52VSVFSTEE439 pKa = 3.87EE440 pKa = 4.17LYY442 pKa = 11.03LAAYY446 pKa = 6.32GTSGAATFGGFYY458 pKa = 10.32SGFTFEE464 pKa = 5.02PEE466 pKa = 3.96VSFTQLDD473 pKa = 4.25LSQANCIPNTSLSVNTLSPFDD494 pKa = 3.54TFQWYY499 pKa = 9.88FNGNIIPSATSSSYY513 pKa = 10.95VPSTLGPGYY522 pKa = 9.81YY523 pKa = 9.39YY524 pKa = 11.24VSATISNCSLPINSNEE540 pKa = 3.89IPVSICSIDD549 pKa = 3.6TDD551 pKa = 4.4NDD553 pKa = 3.64GTNNNIDD560 pKa = 4.48LDD562 pKa = 4.01NDD564 pKa = 3.22NDD566 pKa = 5.37GITNCNEE573 pKa = 3.91SYY575 pKa = 11.31GDD577 pKa = 3.44QGVNLTNTAIGSVNIDD593 pKa = 3.04NYY595 pKa = 9.91TNSFTGSVAFSGTGTPSATPIVGDD619 pKa = 3.61ANGNFVTEE627 pKa = 4.27AANGKK632 pKa = 6.86QNKK635 pKa = 9.33VSFTTSWNNPISLAVEE651 pKa = 4.23YY652 pKa = 8.5ATTALGDD659 pKa = 4.05DD660 pKa = 4.65LFTTSTEE667 pKa = 3.69IRR669 pKa = 11.84ITCPINKK676 pKa = 8.66TLTILNPDD684 pKa = 3.38NQVLVDD690 pKa = 3.68TNYY693 pKa = 11.34DD694 pKa = 3.68GIFEE698 pKa = 4.37SGITEE703 pKa = 3.91YY704 pKa = 11.3SSFEE708 pKa = 3.35IRR710 pKa = 11.84FRR712 pKa = 11.84LNDD715 pKa = 3.71NVPLVAGTGTFSVRR729 pKa = 11.84GNLIDD734 pKa = 5.52AIIVTNVNLVDD745 pKa = 3.98TNTSRR750 pKa = 11.84AAMRR754 pKa = 11.84LIATCVPKK762 pKa = 10.89DD763 pKa = 3.52SDD765 pKa = 3.74NDD767 pKa = 4.03GVPDD771 pKa = 4.3QIDD774 pKa = 3.34WDD776 pKa = 4.28SDD778 pKa = 3.15NDD780 pKa = 4.31TIPDD784 pKa = 4.85FIEE787 pKa = 4.22SQGQNISPLTNVDD800 pKa = 3.77TNNDD804 pKa = 4.13GIDD807 pKa = 3.66DD808 pKa = 4.37LFGAGTTAADD818 pKa = 4.11SDD820 pKa = 4.07TDD822 pKa = 4.59GIPNYY827 pKa = 10.61LDD829 pKa = 4.76LDD831 pKa = 3.96SDD833 pKa = 3.79NDD835 pKa = 4.69GIFDD839 pKa = 4.49IIEE842 pKa = 4.33SGSTGNGSNTNGVTINPVGSNGLDD866 pKa = 3.25NGLEE870 pKa = 4.03TALDD874 pKa = 3.68NGIINYY880 pKa = 8.1TIADD884 pKa = 3.68SDD886 pKa = 3.98ADD888 pKa = 3.69GVLNYY893 pKa = 9.98IEE895 pKa = 5.45IDD897 pKa = 3.92SDD899 pKa = 4.42DD900 pKa = 5.01DD901 pKa = 3.79GCKK904 pKa = 10.38DD905 pKa = 3.27VTEE908 pKa = 5.13AGFTDD913 pKa = 3.81TNNDD917 pKa = 4.1GIVGDD922 pKa = 4.03DD923 pKa = 3.75TPTVVANQGQVISSSGYY940 pKa = 8.82GVPNGDD946 pKa = 3.69YY947 pKa = 10.07TISAPISISAHH958 pKa = 5.41PVDD961 pKa = 3.85VTTCEE966 pKa = 4.07LQSATFTITSVTVDD980 pKa = 3.28SYY982 pKa = 11.22QWQLSTDD989 pKa = 4.6DD990 pKa = 3.7EE991 pKa = 4.83TSWTDD996 pKa = 3.29LANNATYY1003 pKa = 10.86AGVNTVSLTVSNVSPSMVGYY1023 pKa = 9.96KK1024 pKa = 10.32YY1025 pKa = 10.44RR1026 pKa = 11.84VFINKK1031 pKa = 9.25NGNACGLYY1039 pKa = 10.26SEE1041 pKa = 5.62AATLTTFALPVITTPISLIQCDD1063 pKa = 4.79DD1064 pKa = 3.57DD1065 pKa = 3.76TDD1067 pKa = 5.01AISVFNLTQKK1077 pKa = 11.17NDD1079 pKa = 3.89MISANYY1085 pKa = 10.04LNEE1088 pKa = 3.94TFSYY1092 pKa = 8.14FTSSAAANTEE1102 pKa = 3.79NSSFEE1107 pKa = 3.83ITNPLAYY1114 pKa = 9.28TSGNASVYY1122 pKa = 10.87ARR1124 pKa = 11.84VEE1126 pKa = 3.93NSDD1129 pKa = 3.08GCFRR1133 pKa = 11.84VAQINLIVSVTQIPANFVIPNQYY1156 pKa = 10.84LCDD1159 pKa = 5.0DD1160 pKa = 4.33YY1161 pKa = 11.88LDD1163 pKa = 4.31AANDD1167 pKa = 3.7DD1168 pKa = 3.96RR1169 pKa = 11.84DD1170 pKa = 4.55GISGPFNFTAIQNSLAAILPNNTTIKK1196 pKa = 10.24FYY1198 pKa = 9.72KK1199 pKa = 10.03TEE1201 pKa = 4.16ADD1203 pKa = 4.36FLAEE1207 pKa = 4.09TDD1209 pKa = 3.49ASGNPLAIADD1219 pKa = 3.46ITNYY1223 pKa = 10.54RR1224 pKa = 11.84NIGFTNTQKK1233 pKa = 10.03IWVRR1237 pKa = 11.84VDD1239 pKa = 3.14STIDD1243 pKa = 3.27NSCFGFKK1250 pKa = 10.16TFDD1253 pKa = 3.54VVVEE1257 pKa = 4.26ALPVANPMNANNLIRR1272 pKa = 11.84QCDD1275 pKa = 3.86DD1276 pKa = 3.3DD1277 pKa = 5.42HH1278 pKa = 8.0DD1279 pKa = 4.27GTFGFDD1285 pKa = 2.89TSGIQTAVLNGQTGVNVTYY1304 pKa = 10.35FRR1306 pKa = 11.84ADD1308 pKa = 3.14GTQILPFTNPYY1319 pKa = 10.36NVTGTEE1325 pKa = 4.22TITIRR1330 pKa = 11.84VNNNITQTGGQPCYY1344 pKa = 10.85DD1345 pKa = 3.5EE1346 pKa = 5.77VSLQFIVDD1354 pKa = 4.1DD1355 pKa = 4.32LPEE1358 pKa = 4.67AFAVPTTLTTTCDD1371 pKa = 3.57DD1372 pKa = 3.86EE1373 pKa = 6.24ANSIDD1378 pKa = 4.11QDD1380 pKa = 3.88GLFDD1384 pKa = 5.22FDD1386 pKa = 3.66TSTFLSTIIGTQTGMNVYY1404 pKa = 10.45YY1405 pKa = 10.53YY1406 pKa = 10.64DD1407 pKa = 4.21EE1408 pKa = 4.9NNVLIPSSIPNQLPNPFRR1426 pKa = 11.84TATQNVTVVVEE1437 pKa = 3.84NAINNSCSVQVILPFVVNPTPKK1459 pKa = 8.83IDD1461 pKa = 3.57QEE1463 pKa = 4.26EE1464 pKa = 4.32TVIICIPATQVLIDD1478 pKa = 3.88AGIVDD1483 pKa = 4.85GTPTSNYY1490 pKa = 7.99TYY1492 pKa = 9.72QWYY1495 pKa = 9.38FNNVIMPGRR1504 pKa = 11.84TNYY1507 pKa = 10.88SLIASTPGIYY1517 pKa = 10.16SVTVTNSFGCSKK1529 pKa = 9.74TRR1531 pKa = 11.84IITVNGSEE1539 pKa = 4.14IATLQSIDD1547 pKa = 3.82VVDD1550 pKa = 4.87LSDD1553 pKa = 3.55INSISVIVSGSGAYY1567 pKa = 9.18EE1568 pKa = 4.18FSLDD1572 pKa = 4.88DD1573 pKa = 4.42INGPYY1578 pKa = 9.37QDD1580 pKa = 4.53SGLFNNVPMGLHH1592 pKa = 5.2QLYY1595 pKa = 10.25IRR1597 pKa = 11.84DD1598 pKa = 3.86KK1599 pKa = 10.4NGCGSLGPIGIPVLGIPQYY1618 pKa = 8.02FTPNGDD1624 pKa = 3.42GFHH1627 pKa = 7.65DD1628 pKa = 3.75YY1629 pKa = 10.88WNVKK1633 pKa = 8.1GVSMFSNTNSIIFIFDD1649 pKa = 3.4RR1650 pKa = 11.84YY1651 pKa = 10.64GKK1653 pKa = 10.07LLKK1656 pKa = 10.07QLNAFSPGWDD1666 pKa = 2.93GTYY1669 pKa = 10.73NGNQMPADD1677 pKa = 4.6DD1678 pKa = 4.29YY1679 pKa = 10.83WYY1681 pKa = 10.51SIQFDD1686 pKa = 3.53DD1687 pKa = 3.66GRR1689 pKa = 11.84NVKK1692 pKa = 9.68GHH1694 pKa = 6.4FALKK1698 pKa = 10.27RR1699 pKa = 3.54
MM1 pKa = 7.55KK2 pKa = 10.52KK3 pKa = 10.2NLFFLLFIAFLTTNCFSQFSKK24 pKa = 8.35THH26 pKa = 6.21YY27 pKa = 9.98IPPLSGSSNVTAEE40 pKa = 4.07DD41 pKa = 3.28QFIYY45 pKa = 10.54ISTPNINPINFRR57 pKa = 11.84IIEE60 pKa = 4.2LGGSVINGSVSRR72 pKa = 11.84STPFIYY78 pKa = 10.64NIGFGNDD85 pKa = 3.19TQLHH89 pKa = 5.47VDD91 pKa = 3.93ASQTNNTFSNKK102 pKa = 9.49GYY104 pKa = 10.18IIEE107 pKa = 4.9ADD109 pKa = 3.49DD110 pKa = 5.02LIYY113 pKa = 11.18VSARR117 pKa = 11.84VTAGNGNHH125 pKa = 6.81AGEE128 pKa = 4.55LVSKK132 pKa = 10.68GKK134 pKa = 10.6ASLGNRR140 pKa = 11.84FRR142 pKa = 11.84IGAITNLTVDD152 pKa = 4.34NYY154 pKa = 11.39NDD156 pKa = 2.8IHH158 pKa = 5.5TTFISVLASEE168 pKa = 5.02NNTTVNFSDD177 pKa = 4.75FKK179 pKa = 11.12PGVQLINSSTGDD191 pKa = 3.53SPFSIVLNSGEE202 pKa = 4.39SYY204 pKa = 10.89VIAVQGPLNPNRR216 pKa = 11.84DD217 pKa = 3.58GLIGSLVTSDD227 pKa = 4.84KK228 pKa = 10.9PIALNCGSFGGSNAAGNLDD247 pKa = 3.99LGFDD251 pKa = 3.68QIVPAEE257 pKa = 4.17RR258 pKa = 11.84INNNEE263 pKa = 3.96YY264 pKa = 10.54IFIKK268 pKa = 9.68STGVDD273 pKa = 2.85IVEE276 pKa = 4.46TVLLVADD283 pKa = 3.93VDD285 pKa = 3.86NTEE288 pKa = 4.12VFLSGSTTPDD298 pKa = 3.07VILNAGNYY306 pKa = 9.92AVFNGSSFDD315 pKa = 4.23LNGNLYY321 pKa = 9.95IRR323 pKa = 11.84TNQKK327 pKa = 9.19VFAYY331 pKa = 7.92QTIGDD336 pKa = 4.23NSASDD341 pKa = 3.44QRR343 pKa = 11.84NQEE346 pKa = 4.12MFFVPPLSCQTPRR359 pKa = 11.84IIDD362 pKa = 4.21NIPSIDD368 pKa = 3.8YY369 pKa = 10.28VGSRR373 pKa = 11.84QFTGRR378 pKa = 11.84VTIVTKK384 pKa = 10.37TGASLNFIIDD394 pKa = 3.59GTNYY398 pKa = 9.56TLAEE402 pKa = 4.32LSLITNITGPTNVVGNNNYY421 pKa = 6.19QTYY424 pKa = 10.57VITGLSGDD432 pKa = 3.52VSVFSTEE439 pKa = 3.87EE440 pKa = 4.17LYY442 pKa = 11.03LAAYY446 pKa = 6.32GTSGAATFGGFYY458 pKa = 10.32SGFTFEE464 pKa = 5.02PEE466 pKa = 3.96VSFTQLDD473 pKa = 4.25LSQANCIPNTSLSVNTLSPFDD494 pKa = 3.54TFQWYY499 pKa = 9.88FNGNIIPSATSSSYY513 pKa = 10.95VPSTLGPGYY522 pKa = 9.81YY523 pKa = 9.39YY524 pKa = 11.24VSATISNCSLPINSNEE540 pKa = 3.89IPVSICSIDD549 pKa = 3.6TDD551 pKa = 4.4NDD553 pKa = 3.64GTNNNIDD560 pKa = 4.48LDD562 pKa = 4.01NDD564 pKa = 3.22NDD566 pKa = 5.37GITNCNEE573 pKa = 3.91SYY575 pKa = 11.31GDD577 pKa = 3.44QGVNLTNTAIGSVNIDD593 pKa = 3.04NYY595 pKa = 9.91TNSFTGSVAFSGTGTPSATPIVGDD619 pKa = 3.61ANGNFVTEE627 pKa = 4.27AANGKK632 pKa = 6.86QNKK635 pKa = 9.33VSFTTSWNNPISLAVEE651 pKa = 4.23YY652 pKa = 8.5ATTALGDD659 pKa = 4.05DD660 pKa = 4.65LFTTSTEE667 pKa = 3.69IRR669 pKa = 11.84ITCPINKK676 pKa = 8.66TLTILNPDD684 pKa = 3.38NQVLVDD690 pKa = 3.68TNYY693 pKa = 11.34DD694 pKa = 3.68GIFEE698 pKa = 4.37SGITEE703 pKa = 3.91YY704 pKa = 11.3SSFEE708 pKa = 3.35IRR710 pKa = 11.84FRR712 pKa = 11.84LNDD715 pKa = 3.71NVPLVAGTGTFSVRR729 pKa = 11.84GNLIDD734 pKa = 5.52AIIVTNVNLVDD745 pKa = 3.98TNTSRR750 pKa = 11.84AAMRR754 pKa = 11.84LIATCVPKK762 pKa = 10.89DD763 pKa = 3.52SDD765 pKa = 3.74NDD767 pKa = 4.03GVPDD771 pKa = 4.3QIDD774 pKa = 3.34WDD776 pKa = 4.28SDD778 pKa = 3.15NDD780 pKa = 4.31TIPDD784 pKa = 4.85FIEE787 pKa = 4.22SQGQNISPLTNVDD800 pKa = 3.77TNNDD804 pKa = 4.13GIDD807 pKa = 3.66DD808 pKa = 4.37LFGAGTTAADD818 pKa = 4.11SDD820 pKa = 4.07TDD822 pKa = 4.59GIPNYY827 pKa = 10.61LDD829 pKa = 4.76LDD831 pKa = 3.96SDD833 pKa = 3.79NDD835 pKa = 4.69GIFDD839 pKa = 4.49IIEE842 pKa = 4.33SGSTGNGSNTNGVTINPVGSNGLDD866 pKa = 3.25NGLEE870 pKa = 4.03TALDD874 pKa = 3.68NGIINYY880 pKa = 8.1TIADD884 pKa = 3.68SDD886 pKa = 3.98ADD888 pKa = 3.69GVLNYY893 pKa = 9.98IEE895 pKa = 5.45IDD897 pKa = 3.92SDD899 pKa = 4.42DD900 pKa = 5.01DD901 pKa = 3.79GCKK904 pKa = 10.38DD905 pKa = 3.27VTEE908 pKa = 5.13AGFTDD913 pKa = 3.81TNNDD917 pKa = 4.1GIVGDD922 pKa = 4.03DD923 pKa = 3.75TPTVVANQGQVISSSGYY940 pKa = 8.82GVPNGDD946 pKa = 3.69YY947 pKa = 10.07TISAPISISAHH958 pKa = 5.41PVDD961 pKa = 3.85VTTCEE966 pKa = 4.07LQSATFTITSVTVDD980 pKa = 3.28SYY982 pKa = 11.22QWQLSTDD989 pKa = 4.6DD990 pKa = 3.7EE991 pKa = 4.83TSWTDD996 pKa = 3.29LANNATYY1003 pKa = 10.86AGVNTVSLTVSNVSPSMVGYY1023 pKa = 9.96KK1024 pKa = 10.32YY1025 pKa = 10.44RR1026 pKa = 11.84VFINKK1031 pKa = 9.25NGNACGLYY1039 pKa = 10.26SEE1041 pKa = 5.62AATLTTFALPVITTPISLIQCDD1063 pKa = 4.79DD1064 pKa = 3.57DD1065 pKa = 3.76TDD1067 pKa = 5.01AISVFNLTQKK1077 pKa = 11.17NDD1079 pKa = 3.89MISANYY1085 pKa = 10.04LNEE1088 pKa = 3.94TFSYY1092 pKa = 8.14FTSSAAANTEE1102 pKa = 3.79NSSFEE1107 pKa = 3.83ITNPLAYY1114 pKa = 9.28TSGNASVYY1122 pKa = 10.87ARR1124 pKa = 11.84VEE1126 pKa = 3.93NSDD1129 pKa = 3.08GCFRR1133 pKa = 11.84VAQINLIVSVTQIPANFVIPNQYY1156 pKa = 10.84LCDD1159 pKa = 5.0DD1160 pKa = 4.33YY1161 pKa = 11.88LDD1163 pKa = 4.31AANDD1167 pKa = 3.7DD1168 pKa = 3.96RR1169 pKa = 11.84DD1170 pKa = 4.55GISGPFNFTAIQNSLAAILPNNTTIKK1196 pKa = 10.24FYY1198 pKa = 9.72KK1199 pKa = 10.03TEE1201 pKa = 4.16ADD1203 pKa = 4.36FLAEE1207 pKa = 4.09TDD1209 pKa = 3.49ASGNPLAIADD1219 pKa = 3.46ITNYY1223 pKa = 10.54RR1224 pKa = 11.84NIGFTNTQKK1233 pKa = 10.03IWVRR1237 pKa = 11.84VDD1239 pKa = 3.14STIDD1243 pKa = 3.27NSCFGFKK1250 pKa = 10.16TFDD1253 pKa = 3.54VVVEE1257 pKa = 4.26ALPVANPMNANNLIRR1272 pKa = 11.84QCDD1275 pKa = 3.86DD1276 pKa = 3.3DD1277 pKa = 5.42HH1278 pKa = 8.0DD1279 pKa = 4.27GTFGFDD1285 pKa = 2.89TSGIQTAVLNGQTGVNVTYY1304 pKa = 10.35FRR1306 pKa = 11.84ADD1308 pKa = 3.14GTQILPFTNPYY1319 pKa = 10.36NVTGTEE1325 pKa = 4.22TITIRR1330 pKa = 11.84VNNNITQTGGQPCYY1344 pKa = 10.85DD1345 pKa = 3.5EE1346 pKa = 5.77VSLQFIVDD1354 pKa = 4.1DD1355 pKa = 4.32LPEE1358 pKa = 4.67AFAVPTTLTTTCDD1371 pKa = 3.57DD1372 pKa = 3.86EE1373 pKa = 6.24ANSIDD1378 pKa = 4.11QDD1380 pKa = 3.88GLFDD1384 pKa = 5.22FDD1386 pKa = 3.66TSTFLSTIIGTQTGMNVYY1404 pKa = 10.45YY1405 pKa = 10.53YY1406 pKa = 10.64DD1407 pKa = 4.21EE1408 pKa = 4.9NNVLIPSSIPNQLPNPFRR1426 pKa = 11.84TATQNVTVVVEE1437 pKa = 3.84NAINNSCSVQVILPFVVNPTPKK1459 pKa = 8.83IDD1461 pKa = 3.57QEE1463 pKa = 4.26EE1464 pKa = 4.32TVIICIPATQVLIDD1478 pKa = 3.88AGIVDD1483 pKa = 4.85GTPTSNYY1490 pKa = 7.99TYY1492 pKa = 9.72QWYY1495 pKa = 9.38FNNVIMPGRR1504 pKa = 11.84TNYY1507 pKa = 10.88SLIASTPGIYY1517 pKa = 10.16SVTVTNSFGCSKK1529 pKa = 9.74TRR1531 pKa = 11.84IITVNGSEE1539 pKa = 4.14IATLQSIDD1547 pKa = 3.82VVDD1550 pKa = 4.87LSDD1553 pKa = 3.55INSISVIVSGSGAYY1567 pKa = 9.18EE1568 pKa = 4.18FSLDD1572 pKa = 4.88DD1573 pKa = 4.42INGPYY1578 pKa = 9.37QDD1580 pKa = 4.53SGLFNNVPMGLHH1592 pKa = 5.2QLYY1595 pKa = 10.25IRR1597 pKa = 11.84DD1598 pKa = 3.86KK1599 pKa = 10.4NGCGSLGPIGIPVLGIPQYY1618 pKa = 8.02FTPNGDD1624 pKa = 3.42GFHH1627 pKa = 7.65DD1628 pKa = 3.75YY1629 pKa = 10.88WNVKK1633 pKa = 8.1GVSMFSNTNSIIFIFDD1649 pKa = 3.4RR1650 pKa = 11.84YY1651 pKa = 10.64GKK1653 pKa = 10.07LLKK1656 pKa = 10.07QLNAFSPGWDD1666 pKa = 2.93GTYY1669 pKa = 10.73NGNQMPADD1677 pKa = 4.6DD1678 pKa = 4.29YY1679 pKa = 10.83WYY1681 pKa = 10.51SIQFDD1686 pKa = 3.53DD1687 pKa = 3.66GRR1689 pKa = 11.84NVKK1692 pKa = 9.68GHH1694 pKa = 6.4FALKK1698 pKa = 10.27RR1699 pKa = 3.54
Molecular weight: 182.81 kDa
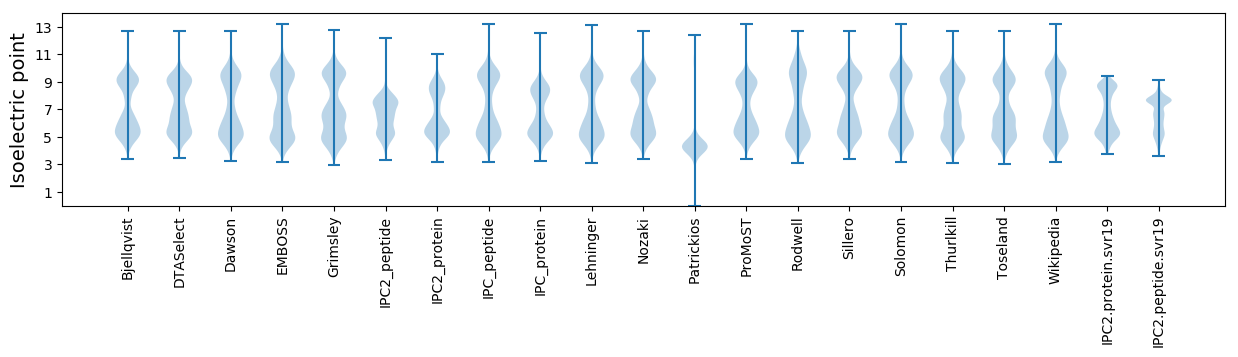
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A495KLC1|A0A495KLC1_9FLAO FkbM family methyltransferase OS=Flavobacterium sp. 102 OX=2135623 GN=C8C84_1577 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1143900 |
29 |
5181 |
344.8 |
38.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.505 ± 0.056 | 0.867 ± 0.028 |
5.249 ± 0.037 | 6.252 ± 0.069 |
5.352 ± 0.04 | 6.198 ± 0.056 |
1.62 ± 0.022 | 8.038 ± 0.047 |
7.677 ± 0.098 | 9.025 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.154 ± 0.027 | 6.59 ± 0.048 |
3.405 ± 0.033 | 3.512 ± 0.025 |
3.061 ± 0.035 | 6.61 ± 0.058 |
6.5 ± 0.118 | 6.312 ± 0.042 |
1.01 ± 0.016 | 4.064 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
