
Lentibacillus salicampi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Lentibacillus
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
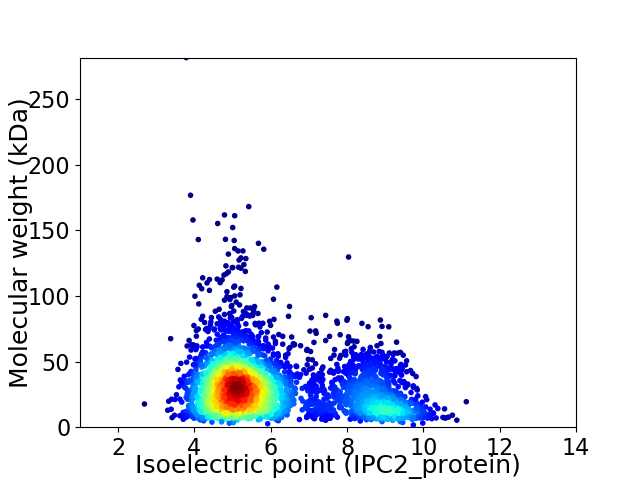
Virtual 2D-PAGE plot for 3721 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9A7L5|A0A4Y9A7L5_9BACI Ectoine/hydroxyectoine ABC transporter permease subunit EhuD OS=Lentibacillus salicampi OX=175306 GN=ehuD PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.52LVGLLLVVLLFALAGCGSAQNEE24 pKa = 4.41GDD26 pKa = 4.29DD27 pKa = 4.31SDD29 pKa = 4.89SGSDD33 pKa = 4.27DD34 pKa = 3.57GGGDD38 pKa = 3.41EE39 pKa = 4.82ASSSEE44 pKa = 4.14EE45 pKa = 3.64VRR47 pKa = 11.84ITLGTGGMSGTYY59 pKa = 9.68YY60 pKa = 10.35PLGVAMTEE68 pKa = 4.23QIFSNAEE75 pKa = 3.73GVSQARR81 pKa = 11.84AVSTGASVTNAQEE94 pKa = 3.93LAEE97 pKa = 4.35GKK99 pKa = 9.2YY100 pKa = 9.23QAVLVQNDD108 pKa = 3.0IAYY111 pKa = 9.62YY112 pKa = 9.94AVNGEE117 pKa = 4.18TLADD121 pKa = 3.95FEE123 pKa = 5.55GNTVDD128 pKa = 4.96NMAGMTSLYY137 pKa = 10.11PEE139 pKa = 5.76DD140 pKa = 3.57IQVVTTADD148 pKa = 3.27SDD150 pKa = 3.97IEE152 pKa = 4.18SLEE155 pKa = 3.98DD156 pKa = 3.52LKK158 pKa = 11.51GKK160 pKa = 9.85NVAVGDD166 pKa = 3.83QGSGAEE172 pKa = 4.08ANANQILEE180 pKa = 4.37AAGITYY186 pKa = 10.78DD187 pKa = 5.12DD188 pKa = 3.43ITAEE192 pKa = 4.03FMGFGDD198 pKa = 5.28ASQGLQNGTIDD209 pKa = 3.38AAFIVAGAPTSAIQEE224 pKa = 4.05LGANKK229 pKa = 9.96DD230 pKa = 3.18IRR232 pKa = 11.84VLSLSDD238 pKa = 4.18EE239 pKa = 4.68IISNLTSEE247 pKa = 4.1YY248 pKa = 10.63SYY250 pKa = 10.13YY251 pKa = 9.68TEE253 pKa = 3.99RR254 pKa = 11.84TIKK257 pKa = 10.48ADD259 pKa = 3.95LYY261 pKa = 10.99ADD263 pKa = 3.63YY264 pKa = 10.49GQEE267 pKa = 3.85EE268 pKa = 5.85DD269 pKa = 3.45ITTVAVMAILVVDD282 pKa = 4.06SEE284 pKa = 4.48LPEE287 pKa = 4.05DD288 pKa = 3.75VVYY291 pKa = 11.56NMTKK295 pKa = 11.02DD296 pKa = 3.09MFEE299 pKa = 4.34NKK301 pKa = 9.75DD302 pKa = 3.6ALEE305 pKa = 4.31AAHH308 pKa = 7.15DD309 pKa = 4.24RR310 pKa = 11.84GSDD313 pKa = 3.13ITLEE317 pKa = 4.12TATDD321 pKa = 3.51GMSIDD326 pKa = 4.16LHH328 pKa = 6.36PGAAKK333 pKa = 10.21YY334 pKa = 10.41YY335 pKa = 8.18EE336 pKa = 4.19EE337 pKa = 4.98EE338 pKa = 4.48GVSVDD343 pKa = 3.35
MM1 pKa = 7.44KK2 pKa = 10.52LVGLLLVVLLFALAGCGSAQNEE24 pKa = 4.41GDD26 pKa = 4.29DD27 pKa = 4.31SDD29 pKa = 4.89SGSDD33 pKa = 4.27DD34 pKa = 3.57GGGDD38 pKa = 3.41EE39 pKa = 4.82ASSSEE44 pKa = 4.14EE45 pKa = 3.64VRR47 pKa = 11.84ITLGTGGMSGTYY59 pKa = 9.68YY60 pKa = 10.35PLGVAMTEE68 pKa = 4.23QIFSNAEE75 pKa = 3.73GVSQARR81 pKa = 11.84AVSTGASVTNAQEE94 pKa = 3.93LAEE97 pKa = 4.35GKK99 pKa = 9.2YY100 pKa = 9.23QAVLVQNDD108 pKa = 3.0IAYY111 pKa = 9.62YY112 pKa = 9.94AVNGEE117 pKa = 4.18TLADD121 pKa = 3.95FEE123 pKa = 5.55GNTVDD128 pKa = 4.96NMAGMTSLYY137 pKa = 10.11PEE139 pKa = 5.76DD140 pKa = 3.57IQVVTTADD148 pKa = 3.27SDD150 pKa = 3.97IEE152 pKa = 4.18SLEE155 pKa = 3.98DD156 pKa = 3.52LKK158 pKa = 11.51GKK160 pKa = 9.85NVAVGDD166 pKa = 3.83QGSGAEE172 pKa = 4.08ANANQILEE180 pKa = 4.37AAGITYY186 pKa = 10.78DD187 pKa = 5.12DD188 pKa = 3.43ITAEE192 pKa = 4.03FMGFGDD198 pKa = 5.28ASQGLQNGTIDD209 pKa = 3.38AAFIVAGAPTSAIQEE224 pKa = 4.05LGANKK229 pKa = 9.96DD230 pKa = 3.18IRR232 pKa = 11.84VLSLSDD238 pKa = 4.18EE239 pKa = 4.68IISNLTSEE247 pKa = 4.1YY248 pKa = 10.63SYY250 pKa = 10.13YY251 pKa = 9.68TEE253 pKa = 3.99RR254 pKa = 11.84TIKK257 pKa = 10.48ADD259 pKa = 3.95LYY261 pKa = 10.99ADD263 pKa = 3.63YY264 pKa = 10.49GQEE267 pKa = 3.85EE268 pKa = 5.85DD269 pKa = 3.45ITTVAVMAILVVDD282 pKa = 4.06SEE284 pKa = 4.48LPEE287 pKa = 4.05DD288 pKa = 3.75VVYY291 pKa = 11.56NMTKK295 pKa = 11.02DD296 pKa = 3.09MFEE299 pKa = 4.34NKK301 pKa = 9.75DD302 pKa = 3.6ALEE305 pKa = 4.31AAHH308 pKa = 7.15DD309 pKa = 4.24RR310 pKa = 11.84GSDD313 pKa = 3.13ITLEE317 pKa = 4.12TATDD321 pKa = 3.51GMSIDD326 pKa = 4.16LHH328 pKa = 6.36PGAAKK333 pKa = 10.21YY334 pKa = 10.41YY335 pKa = 8.18EE336 pKa = 4.19EE337 pKa = 4.98EE338 pKa = 4.48GVSVDD343 pKa = 3.35
Molecular weight: 36.01 kDa
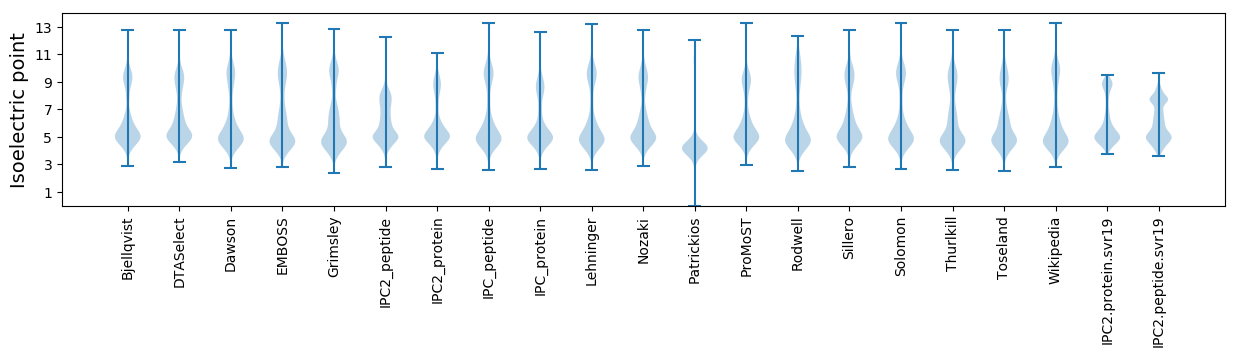
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9ACP1|A0A4Y9ACP1_9BACI Uncharacterized protein OS=Lentibacillus salicampi OX=175306 GN=E4U82_06335 PE=4 SV=1
MM1 pKa = 7.23IRR3 pKa = 11.84SSIYY7 pKa = 10.32ALSLRR12 pKa = 11.84KK13 pKa = 9.82CINRR17 pKa = 11.84ASTAPTRR24 pKa = 11.84ASTTPTRR31 pKa = 11.84ASTAPTRR38 pKa = 11.84ASTTPTRR45 pKa = 11.84ASTAQTRR52 pKa = 11.84ASTTPTRR59 pKa = 11.84ASTTPTRR66 pKa = 11.84ASTTPTRR73 pKa = 11.84ASTTPTRR80 pKa = 11.84ASTTPTRR87 pKa = 11.84ASTAQTRR94 pKa = 11.84ASTVQNRR101 pKa = 11.84ARR103 pKa = 11.84AASTRR108 pKa = 11.84AGTAQTRR115 pKa = 11.84ARR117 pKa = 11.84TAQTRR122 pKa = 11.84AGTALTRR129 pKa = 11.84AGIAQTRR136 pKa = 11.84ARR138 pKa = 11.84TAQTRR143 pKa = 11.84AGTVQTRR150 pKa = 11.84ASTAQTRR157 pKa = 11.84AGTALTRR164 pKa = 11.84AGIAQTRR171 pKa = 11.84AKK173 pKa = 8.98KK174 pKa = 8.87TKK176 pKa = 10.48LRR178 pKa = 11.84DD179 pKa = 3.55APPLQNQQ186 pKa = 3.21
MM1 pKa = 7.23IRR3 pKa = 11.84SSIYY7 pKa = 10.32ALSLRR12 pKa = 11.84KK13 pKa = 9.82CINRR17 pKa = 11.84ASTAPTRR24 pKa = 11.84ASTTPTRR31 pKa = 11.84ASTAPTRR38 pKa = 11.84ASTTPTRR45 pKa = 11.84ASTAQTRR52 pKa = 11.84ASTTPTRR59 pKa = 11.84ASTTPTRR66 pKa = 11.84ASTTPTRR73 pKa = 11.84ASTTPTRR80 pKa = 11.84ASTTPTRR87 pKa = 11.84ASTAQTRR94 pKa = 11.84ASTVQNRR101 pKa = 11.84ARR103 pKa = 11.84AASTRR108 pKa = 11.84AGTAQTRR115 pKa = 11.84ARR117 pKa = 11.84TAQTRR122 pKa = 11.84AGTALTRR129 pKa = 11.84AGIAQTRR136 pKa = 11.84ARR138 pKa = 11.84TAQTRR143 pKa = 11.84AGTVQTRR150 pKa = 11.84ASTAQTRR157 pKa = 11.84AGTALTRR164 pKa = 11.84AGIAQTRR171 pKa = 11.84AKK173 pKa = 8.98KK174 pKa = 8.87TKK176 pKa = 10.48LRR178 pKa = 11.84DD179 pKa = 3.55APPLQNQQ186 pKa = 3.21
Molecular weight: 19.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1050092 |
14 |
2509 |
282.2 |
31.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.24 ± 0.035 | 0.624 ± 0.012 |
5.963 ± 0.043 | 7.402 ± 0.046 |
4.409 ± 0.035 | 6.989 ± 0.035 |
2.205 ± 0.019 | 7.369 ± 0.043 |
6.235 ± 0.036 | 9.317 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.964 ± 0.018 | 4.546 ± 0.031 |
3.608 ± 0.023 | 3.992 ± 0.029 |
4.195 ± 0.034 | 5.889 ± 0.029 |
5.606 ± 0.027 | 6.884 ± 0.03 |
1.051 ± 0.016 | 3.511 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
