
Methanosarcina acetivorans (strain ATCC 35395 / DSM 2834 / JCM 12185 / C2A)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; Methanosarcinaceae; Methanosarcina; Methanosarcina acetivorans
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
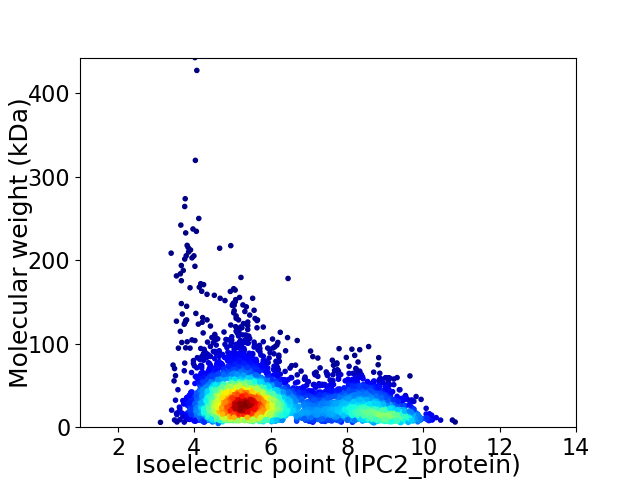
Virtual 2D-PAGE plot for 4468 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8TII5|Q8TII5_METAC Corrinoid protein OS=Methanosarcina acetivorans (strain ATCC 35395 / DSM 2834 / JCM 12185 / C2A) OX=188937 GN=MA_4164 PE=3 SV=1
MM1 pKa = 7.45EE2 pKa = 5.58RR3 pKa = 11.84FNMKK7 pKa = 8.87LTRR10 pKa = 11.84KK11 pKa = 9.82KK12 pKa = 9.84FTGNLLRR19 pKa = 11.84SLAVLLLFSLVVCTAAGEE37 pKa = 4.11EE38 pKa = 5.24LYY40 pKa = 10.81FEE42 pKa = 4.37PQEE45 pKa = 4.55AVFSDD50 pKa = 3.71TGSVASLSLYY60 pKa = 10.47LDD62 pKa = 3.56QAPSGLAGYY71 pKa = 9.64KK72 pKa = 10.39LNLSISDD79 pKa = 3.77PSVAQITGVEE89 pKa = 4.52FPSWSGINNSSSLPADD105 pKa = 3.96LVQIKK110 pKa = 10.35AVDD113 pKa = 3.84LEE115 pKa = 4.3EE116 pKa = 5.16KK117 pKa = 10.57INAGDD122 pKa = 3.95LDD124 pKa = 4.16VFLGTVTVEE133 pKa = 4.11SLASGEE139 pKa = 4.36ANLTVSVDD147 pKa = 3.51RR148 pKa = 11.84LDD150 pKa = 5.06DD151 pKa = 4.42DD152 pKa = 4.25SEE154 pKa = 4.23NTISVTVRR162 pKa = 11.84EE163 pKa = 4.53AKK165 pKa = 9.11LTSGTTSEE173 pKa = 4.38NPVVSILPAEE183 pKa = 4.37STLSSGASQVFEE195 pKa = 3.66IRR197 pKa = 11.84ADD199 pKa = 3.37KK200 pKa = 9.45FTEE203 pKa = 5.06GISGYY208 pKa = 10.7DD209 pKa = 3.41FNLTLEE215 pKa = 4.38NPAVGKK221 pKa = 7.7FTSVEE226 pKa = 4.01YY227 pKa = 9.91PSWVGLSEE235 pKa = 4.06NSSMPNTSIKK245 pKa = 10.49VKK247 pKa = 10.67AVDD250 pKa = 3.49TNDD253 pKa = 3.03VVKK256 pKa = 11.07AGDD259 pKa = 3.96EE260 pKa = 4.18NVLLANVTFVAEE272 pKa = 4.4TPGEE276 pKa = 4.23SEE278 pKa = 4.29ISLTFNRR285 pKa = 11.84LDD287 pKa = 4.57DD288 pKa = 5.0DD289 pKa = 4.09SGSNINAATEE299 pKa = 3.85ASSIKK304 pKa = 10.18VQGSSTLPVAKK315 pKa = 9.39FTATPTSGDD324 pKa = 3.37SPLTVQFTDD333 pKa = 3.65EE334 pKa = 4.5STGSPTAWAWDD345 pKa = 3.58FDD347 pKa = 3.88NDD349 pKa = 3.97GNVDD353 pKa = 3.53STEE356 pKa = 3.96QNPSYY361 pKa = 9.86TYY363 pKa = 10.89SDD365 pKa = 3.0AGIYY369 pKa = 7.29TVKK372 pKa = 10.01LTVSNEE378 pKa = 3.76NGSDD382 pKa = 3.42VLEE385 pKa = 4.24ILEE388 pKa = 4.63MITVKK393 pKa = 9.6EE394 pKa = 4.33TVVSEE399 pKa = 4.07DD400 pKa = 2.44AWYY403 pKa = 10.4QFHH406 pKa = 7.95KK407 pKa = 10.6DD408 pKa = 3.68AQHH411 pKa = 6.21SGYY414 pKa = 10.25SSSDD418 pKa = 3.36APDD421 pKa = 3.54SANLAWIAEE430 pKa = 4.36PLNNTYY436 pKa = 11.18SLVPSSSVVIAEE448 pKa = 3.76GMVFGLCNGPVDD460 pKa = 4.47EE461 pKa = 5.13YY462 pKa = 11.62GNPLTSEE469 pKa = 4.28GQLVAFDD476 pKa = 4.38EE477 pKa = 4.63EE478 pKa = 4.49TGEE481 pKa = 4.36EE482 pKa = 3.87IWNVTVMAPEE492 pKa = 4.18WGSWSCPAYY501 pKa = 10.52DD502 pKa = 4.28DD503 pKa = 4.46GKK505 pKa = 11.12VFASAGKK512 pKa = 8.23NTYY515 pKa = 9.56CVNASTGDD523 pKa = 4.0IIWTFQNPSEE533 pKa = 4.11LASCNGGPSIGDD545 pKa = 3.32GKK547 pKa = 11.05VFASDD552 pKa = 3.14WDD554 pKa = 3.72GGNYY558 pKa = 9.61YY559 pKa = 10.69CLDD562 pKa = 3.88EE563 pKa = 4.38NTGEE567 pKa = 4.31LLWTFKK573 pKa = 10.52IDD575 pKa = 3.34GLYY578 pKa = 10.5AQSTPAYY585 pKa = 9.06KK586 pKa = 10.01DD587 pKa = 3.13DD588 pKa = 3.61RR589 pKa = 11.84VYY591 pKa = 11.31LSGWTTVNAVYY602 pKa = 10.19CVNATTGEE610 pKa = 4.42LIWEE614 pKa = 4.14NEE616 pKa = 4.06EE617 pKa = 5.08LSSNPCGSITVTEE630 pKa = 4.14EE631 pKa = 3.55GLYY634 pKa = 10.7LSIYY638 pKa = 10.78SFGTEE643 pKa = 3.7DD644 pKa = 4.02GFYY647 pKa = 10.95KK648 pKa = 10.66LDD650 pKa = 3.54LTDD653 pKa = 3.63GHH655 pKa = 6.55EE656 pKa = 3.74IWGRR660 pKa = 11.84PDD662 pKa = 3.15IPPTDD667 pKa = 3.61STPAVVNGKK676 pKa = 9.95VYY678 pKa = 10.73LSAGTAGYY686 pKa = 10.65SDD688 pKa = 5.15LNTYY692 pKa = 9.69CLNASDD698 pKa = 5.71GKK700 pKa = 8.81TIWVTDD706 pKa = 3.42SSEE709 pKa = 4.66NIGDD713 pKa = 4.16WVCSPAVADD722 pKa = 4.09GKK724 pKa = 11.11VFTGGAAEE732 pKa = 4.22GLFTGSSTLYY742 pKa = 11.0AFDD745 pKa = 4.96AEE747 pKa = 4.4TGAVIWSYY755 pKa = 10.61KK756 pKa = 9.57GCGGSPAVADD766 pKa = 4.07DD767 pKa = 4.17MVFSTGSGKK776 pKa = 10.73LYY778 pKa = 10.35AFKK781 pKa = 10.26EE782 pKa = 4.34AEE784 pKa = 4.19VLLPEE789 pKa = 4.96AKK791 pKa = 10.07FSSNVSSGEE800 pKa = 3.81APLTVGFTDD809 pKa = 3.45EE810 pKa = 4.4STGEE814 pKa = 4.87GITAWEE820 pKa = 3.95WDD822 pKa = 3.46FDD824 pKa = 3.86NDD826 pKa = 3.98GNVDD830 pKa = 3.55STEE833 pKa = 3.89QNPVHH838 pKa = 6.46TYY840 pKa = 11.42DD841 pKa = 3.18NAGSYY846 pKa = 7.95TVNLTVTSAEE856 pKa = 4.25GSDD859 pKa = 4.01SEE861 pKa = 5.52VKK863 pKa = 9.5MDD865 pKa = 4.24YY866 pKa = 10.95INVSEE871 pKa = 4.51SSTPGEE877 pKa = 4.32PEE879 pKa = 3.49PVAAFIANVTSGTAPLTVNFTDD901 pKa = 3.66QSTGSPTSWEE911 pKa = 3.65WDD913 pKa = 3.09FDD915 pKa = 3.72NDD917 pKa = 3.98GNVDD921 pKa = 3.55STEE924 pKa = 3.89QNPVHH929 pKa = 6.94TYY931 pKa = 8.49TAAGNYY937 pKa = 7.27TVSLTVTNAEE947 pKa = 4.26GSDD950 pKa = 3.81SEE952 pKa = 4.85VKK954 pKa = 10.45ADD956 pKa = 3.84YY957 pKa = 9.24ITVSEE962 pKa = 4.38ASTPVEE968 pKa = 4.27PEE970 pKa = 3.4PVAAFTADD978 pKa = 3.7VTSGTAPLTVNFTDD992 pKa = 3.66QSTGSPTAWAWDD1004 pKa = 3.58FDD1006 pKa = 3.88NDD1008 pKa = 3.97GNVDD1012 pKa = 3.53STEE1015 pKa = 3.96QNPSYY1020 pKa = 10.15TYY1022 pKa = 11.23NDD1024 pKa = 3.46AGSYY1028 pKa = 8.11TVNLTVTNAEE1038 pKa = 4.12GSNSEE1043 pKa = 4.22VKK1045 pKa = 9.78TEE1047 pKa = 4.16YY1048 pKa = 9.41ITVEE1052 pKa = 4.16EE1053 pKa = 4.59ASSGSQNGSSSVSLNVTIVPVISLEE1078 pKa = 4.14VSPSALDD1085 pKa = 3.64FGEE1088 pKa = 4.85LYY1090 pKa = 10.1PGKK1093 pKa = 9.05TSEE1096 pKa = 4.51LQYY1099 pKa = 10.48LTLKK1103 pKa = 10.63NRR1105 pKa = 11.84GSCDD1109 pKa = 3.14INVTAVVCDD1118 pKa = 4.63CSAEE1122 pKa = 4.34DD1123 pKa = 3.66EE1124 pKa = 4.78LFSQGLLLDD1133 pKa = 3.76SQLWNNYY1140 pKa = 6.04WKK1142 pKa = 10.9VVGKK1146 pKa = 10.21NSQEE1150 pKa = 4.02NTSVALQVPADD1161 pKa = 3.82YY1162 pKa = 10.99AGSGNKK1168 pKa = 9.51KK1169 pKa = 10.42GSITFWAEE1177 pKa = 3.08AAEE1180 pKa = 4.12
MM1 pKa = 7.45EE2 pKa = 5.58RR3 pKa = 11.84FNMKK7 pKa = 8.87LTRR10 pKa = 11.84KK11 pKa = 9.82KK12 pKa = 9.84FTGNLLRR19 pKa = 11.84SLAVLLLFSLVVCTAAGEE37 pKa = 4.11EE38 pKa = 5.24LYY40 pKa = 10.81FEE42 pKa = 4.37PQEE45 pKa = 4.55AVFSDD50 pKa = 3.71TGSVASLSLYY60 pKa = 10.47LDD62 pKa = 3.56QAPSGLAGYY71 pKa = 9.64KK72 pKa = 10.39LNLSISDD79 pKa = 3.77PSVAQITGVEE89 pKa = 4.52FPSWSGINNSSSLPADD105 pKa = 3.96LVQIKK110 pKa = 10.35AVDD113 pKa = 3.84LEE115 pKa = 4.3EE116 pKa = 5.16KK117 pKa = 10.57INAGDD122 pKa = 3.95LDD124 pKa = 4.16VFLGTVTVEE133 pKa = 4.11SLASGEE139 pKa = 4.36ANLTVSVDD147 pKa = 3.51RR148 pKa = 11.84LDD150 pKa = 5.06DD151 pKa = 4.42DD152 pKa = 4.25SEE154 pKa = 4.23NTISVTVRR162 pKa = 11.84EE163 pKa = 4.53AKK165 pKa = 9.11LTSGTTSEE173 pKa = 4.38NPVVSILPAEE183 pKa = 4.37STLSSGASQVFEE195 pKa = 3.66IRR197 pKa = 11.84ADD199 pKa = 3.37KK200 pKa = 9.45FTEE203 pKa = 5.06GISGYY208 pKa = 10.7DD209 pKa = 3.41FNLTLEE215 pKa = 4.38NPAVGKK221 pKa = 7.7FTSVEE226 pKa = 4.01YY227 pKa = 9.91PSWVGLSEE235 pKa = 4.06NSSMPNTSIKK245 pKa = 10.49VKK247 pKa = 10.67AVDD250 pKa = 3.49TNDD253 pKa = 3.03VVKK256 pKa = 11.07AGDD259 pKa = 3.96EE260 pKa = 4.18NVLLANVTFVAEE272 pKa = 4.4TPGEE276 pKa = 4.23SEE278 pKa = 4.29ISLTFNRR285 pKa = 11.84LDD287 pKa = 4.57DD288 pKa = 5.0DD289 pKa = 4.09SGSNINAATEE299 pKa = 3.85ASSIKK304 pKa = 10.18VQGSSTLPVAKK315 pKa = 9.39FTATPTSGDD324 pKa = 3.37SPLTVQFTDD333 pKa = 3.65EE334 pKa = 4.5STGSPTAWAWDD345 pKa = 3.58FDD347 pKa = 3.88NDD349 pKa = 3.97GNVDD353 pKa = 3.53STEE356 pKa = 3.96QNPSYY361 pKa = 9.86TYY363 pKa = 10.89SDD365 pKa = 3.0AGIYY369 pKa = 7.29TVKK372 pKa = 10.01LTVSNEE378 pKa = 3.76NGSDD382 pKa = 3.42VLEE385 pKa = 4.24ILEE388 pKa = 4.63MITVKK393 pKa = 9.6EE394 pKa = 4.33TVVSEE399 pKa = 4.07DD400 pKa = 2.44AWYY403 pKa = 10.4QFHH406 pKa = 7.95KK407 pKa = 10.6DD408 pKa = 3.68AQHH411 pKa = 6.21SGYY414 pKa = 10.25SSSDD418 pKa = 3.36APDD421 pKa = 3.54SANLAWIAEE430 pKa = 4.36PLNNTYY436 pKa = 11.18SLVPSSSVVIAEE448 pKa = 3.76GMVFGLCNGPVDD460 pKa = 4.47EE461 pKa = 5.13YY462 pKa = 11.62GNPLTSEE469 pKa = 4.28GQLVAFDD476 pKa = 4.38EE477 pKa = 4.63EE478 pKa = 4.49TGEE481 pKa = 4.36EE482 pKa = 3.87IWNVTVMAPEE492 pKa = 4.18WGSWSCPAYY501 pKa = 10.52DD502 pKa = 4.28DD503 pKa = 4.46GKK505 pKa = 11.12VFASAGKK512 pKa = 8.23NTYY515 pKa = 9.56CVNASTGDD523 pKa = 4.0IIWTFQNPSEE533 pKa = 4.11LASCNGGPSIGDD545 pKa = 3.32GKK547 pKa = 11.05VFASDD552 pKa = 3.14WDD554 pKa = 3.72GGNYY558 pKa = 9.61YY559 pKa = 10.69CLDD562 pKa = 3.88EE563 pKa = 4.38NTGEE567 pKa = 4.31LLWTFKK573 pKa = 10.52IDD575 pKa = 3.34GLYY578 pKa = 10.5AQSTPAYY585 pKa = 9.06KK586 pKa = 10.01DD587 pKa = 3.13DD588 pKa = 3.61RR589 pKa = 11.84VYY591 pKa = 11.31LSGWTTVNAVYY602 pKa = 10.19CVNATTGEE610 pKa = 4.42LIWEE614 pKa = 4.14NEE616 pKa = 4.06EE617 pKa = 5.08LSSNPCGSITVTEE630 pKa = 4.14EE631 pKa = 3.55GLYY634 pKa = 10.7LSIYY638 pKa = 10.78SFGTEE643 pKa = 3.7DD644 pKa = 4.02GFYY647 pKa = 10.95KK648 pKa = 10.66LDD650 pKa = 3.54LTDD653 pKa = 3.63GHH655 pKa = 6.55EE656 pKa = 3.74IWGRR660 pKa = 11.84PDD662 pKa = 3.15IPPTDD667 pKa = 3.61STPAVVNGKK676 pKa = 9.95VYY678 pKa = 10.73LSAGTAGYY686 pKa = 10.65SDD688 pKa = 5.15LNTYY692 pKa = 9.69CLNASDD698 pKa = 5.71GKK700 pKa = 8.81TIWVTDD706 pKa = 3.42SSEE709 pKa = 4.66NIGDD713 pKa = 4.16WVCSPAVADD722 pKa = 4.09GKK724 pKa = 11.11VFTGGAAEE732 pKa = 4.22GLFTGSSTLYY742 pKa = 11.0AFDD745 pKa = 4.96AEE747 pKa = 4.4TGAVIWSYY755 pKa = 10.61KK756 pKa = 9.57GCGGSPAVADD766 pKa = 4.07DD767 pKa = 4.17MVFSTGSGKK776 pKa = 10.73LYY778 pKa = 10.35AFKK781 pKa = 10.26EE782 pKa = 4.34AEE784 pKa = 4.19VLLPEE789 pKa = 4.96AKK791 pKa = 10.07FSSNVSSGEE800 pKa = 3.81APLTVGFTDD809 pKa = 3.45EE810 pKa = 4.4STGEE814 pKa = 4.87GITAWEE820 pKa = 3.95WDD822 pKa = 3.46FDD824 pKa = 3.86NDD826 pKa = 3.98GNVDD830 pKa = 3.55STEE833 pKa = 3.89QNPVHH838 pKa = 6.46TYY840 pKa = 11.42DD841 pKa = 3.18NAGSYY846 pKa = 7.95TVNLTVTSAEE856 pKa = 4.25GSDD859 pKa = 4.01SEE861 pKa = 5.52VKK863 pKa = 9.5MDD865 pKa = 4.24YY866 pKa = 10.95INVSEE871 pKa = 4.51SSTPGEE877 pKa = 4.32PEE879 pKa = 3.49PVAAFIANVTSGTAPLTVNFTDD901 pKa = 3.66QSTGSPTSWEE911 pKa = 3.65WDD913 pKa = 3.09FDD915 pKa = 3.72NDD917 pKa = 3.98GNVDD921 pKa = 3.55STEE924 pKa = 3.89QNPVHH929 pKa = 6.94TYY931 pKa = 8.49TAAGNYY937 pKa = 7.27TVSLTVTNAEE947 pKa = 4.26GSDD950 pKa = 3.81SEE952 pKa = 4.85VKK954 pKa = 10.45ADD956 pKa = 3.84YY957 pKa = 9.24ITVSEE962 pKa = 4.38ASTPVEE968 pKa = 4.27PEE970 pKa = 3.4PVAAFTADD978 pKa = 3.7VTSGTAPLTVNFTDD992 pKa = 3.66QSTGSPTAWAWDD1004 pKa = 3.58FDD1006 pKa = 3.88NDD1008 pKa = 3.97GNVDD1012 pKa = 3.53STEE1015 pKa = 3.96QNPSYY1020 pKa = 10.15TYY1022 pKa = 11.23NDD1024 pKa = 3.46AGSYY1028 pKa = 8.11TVNLTVTNAEE1038 pKa = 4.12GSNSEE1043 pKa = 4.22VKK1045 pKa = 9.78TEE1047 pKa = 4.16YY1048 pKa = 9.41ITVEE1052 pKa = 4.16EE1053 pKa = 4.59ASSGSQNGSSSVSLNVTIVPVISLEE1078 pKa = 4.14VSPSALDD1085 pKa = 3.64FGEE1088 pKa = 4.85LYY1090 pKa = 10.1PGKK1093 pKa = 9.05TSEE1096 pKa = 4.51LQYY1099 pKa = 10.48LTLKK1103 pKa = 10.63NRR1105 pKa = 11.84GSCDD1109 pKa = 3.14INVTAVVCDD1118 pKa = 4.63CSAEE1122 pKa = 4.34DD1123 pKa = 3.66EE1124 pKa = 4.78LFSQGLLLDD1133 pKa = 3.76SQLWNNYY1140 pKa = 6.04WKK1142 pKa = 10.9VVGKK1146 pKa = 10.21NSQEE1150 pKa = 4.02NTSVALQVPADD1161 pKa = 3.82YY1162 pKa = 10.99AGSGNKK1168 pKa = 9.51KK1169 pKa = 10.42GSITFWAEE1177 pKa = 3.08AAEE1180 pKa = 4.12
Molecular weight: 125.54 kDa
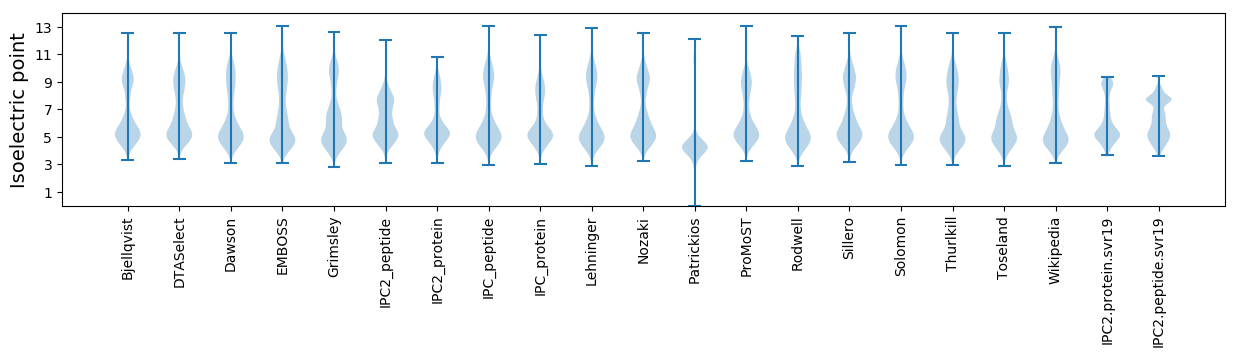
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8TIW5|Q8TIW5_METAC Uncharacterized protein OS=Methanosarcina acetivorans (strain ATCC 35395 / DSM 2834 / JCM 12185 / C2A) OX=188937 GN=MA_4025 PE=4 SV=1
MM1 pKa = 6.8SHH3 pKa = 6.05NMKK6 pKa = 10.11GQKK9 pKa = 9.42KK10 pKa = 9.38RR11 pKa = 11.84LAKK14 pKa = 10.01AHH16 pKa = 5.8KK17 pKa = 9.35QNTRR21 pKa = 11.84VPVWVIVKK29 pKa = 7.36TNRR32 pKa = 11.84KK33 pKa = 8.65VVSHH37 pKa = 6.09PRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.43WRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84SLDD49 pKa = 3.3VKK51 pKa = 11.01
MM1 pKa = 6.8SHH3 pKa = 6.05NMKK6 pKa = 10.11GQKK9 pKa = 9.42KK10 pKa = 9.38RR11 pKa = 11.84LAKK14 pKa = 10.01AHH16 pKa = 5.8KK17 pKa = 9.35QNTRR21 pKa = 11.84VPVWVIVKK29 pKa = 7.36TNRR32 pKa = 11.84KK33 pKa = 8.65VVSHH37 pKa = 6.09PRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.43WRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84SLDD49 pKa = 3.3VKK51 pKa = 11.01
Molecular weight: 6.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1392316 |
41 |
4226 |
311.6 |
34.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.926 ± 0.044 | 1.251 ± 0.018 |
5.35 ± 0.026 | 7.966 ± 0.048 |
4.434 ± 0.031 | 7.263 ± 0.041 |
1.659 ± 0.015 | 7.35 ± 0.038 |
6.498 ± 0.052 | 9.406 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.459 ± 0.021 | 4.474 ± 0.038 |
4.006 ± 0.023 | 2.533 ± 0.019 |
4.465 ± 0.039 | 6.907 ± 0.042 |
5.442 ± 0.049 | 6.843 ± 0.034 |
1.052 ± 0.013 | 3.718 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
