
Cellulophaga lytica (strain ATCC 23178 / DSM 7489 / JCM 8516 / NBRC 14961 / NCIMB 1423 / VKM B-1433 / Cy l20)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Cellulophaga; Cellulophaga lytica
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
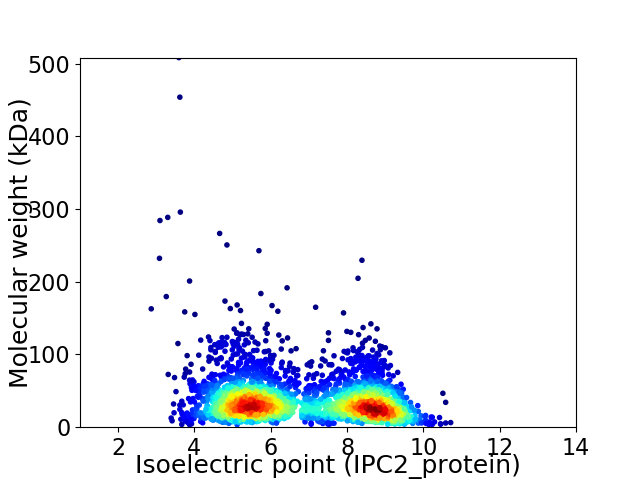
Virtual 2D-PAGE plot for 3281 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0RCN7|F0RCN7_CELLC Peptidase S41 OS=Cellulophaga lytica (strain ATCC 23178 / DSM 7489 / JCM 8516 / NBRC 14961 / NCIMB 1423 / VKM B-1433 / Cy l20) OX=867900 GN=Celly_1911 PE=4 SV=1
MM1 pKa = 7.82KK2 pKa = 10.05IKK4 pKa = 10.5QLGSFKK10 pKa = 10.46CIKK13 pKa = 9.76KK14 pKa = 9.8QRR16 pKa = 11.84TLSLLVMFFCFSFFAFGQNVSVQRR40 pKa = 11.84TNDD43 pKa = 3.36AEE45 pKa = 4.36EE46 pKa = 4.27TGQVAGSFRR55 pKa = 11.84ISRR58 pKa = 11.84TGNTFTGLRR67 pKa = 11.84VNYY70 pKa = 8.61TITGTATNGTDD81 pKa = 3.38YY82 pKa = 11.61ASIGGSILIPVFNNSINLDD101 pKa = 3.04IDD103 pKa = 3.98GVVDD107 pKa = 3.5DD108 pKa = 4.93TLIEE112 pKa = 4.22GDD114 pKa = 3.59EE115 pKa = 4.36TVIITLSEE123 pKa = 3.72NSEE126 pKa = 4.27YY127 pKa = 11.05NINPLAASARR137 pKa = 11.84IIIVDD142 pKa = 3.95NEE144 pKa = 4.47PVCATAPDD152 pKa = 3.95APEE155 pKa = 4.02LRR157 pKa = 11.84TGEE160 pKa = 4.15EE161 pKa = 4.11TTFCDD166 pKa = 5.51SFTKK170 pKa = 10.62DD171 pKa = 3.11LNDD174 pKa = 3.67YY175 pKa = 9.43VASATPAGTVLTWSSSDD192 pKa = 4.22DD193 pKa = 3.85LLDD196 pKa = 4.3DD197 pKa = 3.53SSYY200 pKa = 11.06YY201 pKa = 10.55DD202 pKa = 3.19SSIIGFSGEE211 pKa = 3.95FYY213 pKa = 11.11GFYY216 pKa = 10.5LDD218 pKa = 4.07EE219 pKa = 5.25ANNCVSPPLKK229 pKa = 9.62ITIVQNSSPTVTGTSDD245 pKa = 3.3SRR247 pKa = 11.84CGEE250 pKa = 3.99GSVTLTGTTTDD261 pKa = 5.67GSLLWYY267 pKa = 8.08DD268 pKa = 3.89APSGGTLVHH277 pKa = 6.66TGLNFDD283 pKa = 4.13TPSLSVTTSYY293 pKa = 11.18YY294 pKa = 10.87VEE296 pKa = 4.17ATANGCTSDD305 pKa = 3.42RR306 pKa = 11.84VEE308 pKa = 4.29VVATINTQPSSGTGTDD324 pKa = 3.19ATACSLPGNGRR335 pKa = 11.84VTVVNLDD342 pKa = 3.81DD343 pKa = 3.98QLTGADD349 pKa = 3.39TGEE352 pKa = 4.17WTLTSQPASSSITIDD367 pKa = 3.28ANSVDD372 pKa = 4.96FANQPLGEE380 pKa = 4.15YY381 pKa = 10.1QFTYY385 pKa = 7.56TTTGFVAPCTAEE397 pKa = 3.91STTITITVLDD407 pKa = 4.56CAPTDD412 pKa = 3.48TDD414 pKa = 4.16LSVTKK419 pKa = 10.37IVDD422 pKa = 3.32RR423 pKa = 11.84DD424 pKa = 3.66EE425 pKa = 4.7VFTGEE430 pKa = 4.53DD431 pKa = 3.57VVFTITVEE439 pKa = 4.04NLTGDD444 pKa = 3.75PVTAIEE450 pKa = 4.25ITDD453 pKa = 3.74VLDD456 pKa = 3.39EE457 pKa = 4.48TTGFEE462 pKa = 4.41YY463 pKa = 10.91VSNIAQLGSFNSATGIWSIAEE484 pKa = 3.99LAAGEE489 pKa = 4.42STSLEE494 pKa = 3.68ITATVIDD501 pKa = 4.8GGTHH505 pKa = 6.23INTAEE510 pKa = 4.06ITSSNPVDD518 pKa = 3.13TATANNQSSATVAIKK533 pKa = 10.45QIDD536 pKa = 3.67MSISKK541 pKa = 10.16SVNKK545 pKa = 8.68VTATVGDD552 pKa = 3.8VVVFTITLTNEE563 pKa = 3.68TDD565 pKa = 3.67GLVTDD570 pKa = 4.81ISVTDD575 pKa = 3.85ILNSTTGFTHH585 pKa = 6.77VSNTTTTGNYY595 pKa = 8.91DD596 pKa = 2.99ATTGVWTLDD605 pKa = 3.53EE606 pKa = 4.19ILGNEE611 pKa = 4.24TQVLEE616 pKa = 4.08ITAMVLEE623 pKa = 5.09EE624 pKa = 4.32GTHH627 pKa = 5.31VNTVEE632 pKa = 4.14ITDD635 pKa = 3.93SFPIDD640 pKa = 3.62EE641 pKa = 5.12ANVDD645 pKa = 3.63NNIASVAVSIEE656 pKa = 3.58PRR658 pKa = 11.84SNDD661 pKa = 2.63EE662 pKa = 4.1CGFMFNQFSPNSDD675 pKa = 2.88GVNDD679 pKa = 4.29FLRR682 pKa = 11.84INCIEE687 pKa = 4.18TYY689 pKa = 10.3PNNTLTIFNRR699 pKa = 11.84LGSEE703 pKa = 4.48VYY705 pKa = 10.68SVTNYY710 pKa = 9.46QNNWDD715 pKa = 4.1GTWKK719 pKa = 10.74KK720 pKa = 11.07GDD722 pKa = 4.39LPNGTYY728 pKa = 10.09FYY730 pKa = 11.46VLDD733 pKa = 4.46LADD736 pKa = 3.86GSEE739 pKa = 4.29VKK741 pKa = 10.59KK742 pKa = 10.93GWIQIIRR749 pKa = 3.64
MM1 pKa = 7.82KK2 pKa = 10.05IKK4 pKa = 10.5QLGSFKK10 pKa = 10.46CIKK13 pKa = 9.76KK14 pKa = 9.8QRR16 pKa = 11.84TLSLLVMFFCFSFFAFGQNVSVQRR40 pKa = 11.84TNDD43 pKa = 3.36AEE45 pKa = 4.36EE46 pKa = 4.27TGQVAGSFRR55 pKa = 11.84ISRR58 pKa = 11.84TGNTFTGLRR67 pKa = 11.84VNYY70 pKa = 8.61TITGTATNGTDD81 pKa = 3.38YY82 pKa = 11.61ASIGGSILIPVFNNSINLDD101 pKa = 3.04IDD103 pKa = 3.98GVVDD107 pKa = 3.5DD108 pKa = 4.93TLIEE112 pKa = 4.22GDD114 pKa = 3.59EE115 pKa = 4.36TVIITLSEE123 pKa = 3.72NSEE126 pKa = 4.27YY127 pKa = 11.05NINPLAASARR137 pKa = 11.84IIIVDD142 pKa = 3.95NEE144 pKa = 4.47PVCATAPDD152 pKa = 3.95APEE155 pKa = 4.02LRR157 pKa = 11.84TGEE160 pKa = 4.15EE161 pKa = 4.11TTFCDD166 pKa = 5.51SFTKK170 pKa = 10.62DD171 pKa = 3.11LNDD174 pKa = 3.67YY175 pKa = 9.43VASATPAGTVLTWSSSDD192 pKa = 4.22DD193 pKa = 3.85LLDD196 pKa = 4.3DD197 pKa = 3.53SSYY200 pKa = 11.06YY201 pKa = 10.55DD202 pKa = 3.19SSIIGFSGEE211 pKa = 3.95FYY213 pKa = 11.11GFYY216 pKa = 10.5LDD218 pKa = 4.07EE219 pKa = 5.25ANNCVSPPLKK229 pKa = 9.62ITIVQNSSPTVTGTSDD245 pKa = 3.3SRR247 pKa = 11.84CGEE250 pKa = 3.99GSVTLTGTTTDD261 pKa = 5.67GSLLWYY267 pKa = 8.08DD268 pKa = 3.89APSGGTLVHH277 pKa = 6.66TGLNFDD283 pKa = 4.13TPSLSVTTSYY293 pKa = 11.18YY294 pKa = 10.87VEE296 pKa = 4.17ATANGCTSDD305 pKa = 3.42RR306 pKa = 11.84VEE308 pKa = 4.29VVATINTQPSSGTGTDD324 pKa = 3.19ATACSLPGNGRR335 pKa = 11.84VTVVNLDD342 pKa = 3.81DD343 pKa = 3.98QLTGADD349 pKa = 3.39TGEE352 pKa = 4.17WTLTSQPASSSITIDD367 pKa = 3.28ANSVDD372 pKa = 4.96FANQPLGEE380 pKa = 4.15YY381 pKa = 10.1QFTYY385 pKa = 7.56TTTGFVAPCTAEE397 pKa = 3.91STTITITVLDD407 pKa = 4.56CAPTDD412 pKa = 3.48TDD414 pKa = 4.16LSVTKK419 pKa = 10.37IVDD422 pKa = 3.32RR423 pKa = 11.84DD424 pKa = 3.66EE425 pKa = 4.7VFTGEE430 pKa = 4.53DD431 pKa = 3.57VVFTITVEE439 pKa = 4.04NLTGDD444 pKa = 3.75PVTAIEE450 pKa = 4.25ITDD453 pKa = 3.74VLDD456 pKa = 3.39EE457 pKa = 4.48TTGFEE462 pKa = 4.41YY463 pKa = 10.91VSNIAQLGSFNSATGIWSIAEE484 pKa = 3.99LAAGEE489 pKa = 4.42STSLEE494 pKa = 3.68ITATVIDD501 pKa = 4.8GGTHH505 pKa = 6.23INTAEE510 pKa = 4.06ITSSNPVDD518 pKa = 3.13TATANNQSSATVAIKK533 pKa = 10.45QIDD536 pKa = 3.67MSISKK541 pKa = 10.16SVNKK545 pKa = 8.68VTATVGDD552 pKa = 3.8VVVFTITLTNEE563 pKa = 3.68TDD565 pKa = 3.67GLVTDD570 pKa = 4.81ISVTDD575 pKa = 3.85ILNSTTGFTHH585 pKa = 6.77VSNTTTTGNYY595 pKa = 8.91DD596 pKa = 2.99ATTGVWTLDD605 pKa = 3.53EE606 pKa = 4.19ILGNEE611 pKa = 4.24TQVLEE616 pKa = 4.08ITAMVLEE623 pKa = 5.09EE624 pKa = 4.32GTHH627 pKa = 5.31VNTVEE632 pKa = 4.14ITDD635 pKa = 3.93SFPIDD640 pKa = 3.62EE641 pKa = 5.12ANVDD645 pKa = 3.63NNIASVAVSIEE656 pKa = 3.58PRR658 pKa = 11.84SNDD661 pKa = 2.63EE662 pKa = 4.1CGFMFNQFSPNSDD675 pKa = 2.88GVNDD679 pKa = 4.29FLRR682 pKa = 11.84INCIEE687 pKa = 4.18TYY689 pKa = 10.3PNNTLTIFNRR699 pKa = 11.84LGSEE703 pKa = 4.48VYY705 pKa = 10.68SVTNYY710 pKa = 9.46QNNWDD715 pKa = 4.1GTWKK719 pKa = 10.74KK720 pKa = 11.07GDD722 pKa = 4.39LPNGTYY728 pKa = 10.09FYY730 pKa = 11.46VLDD733 pKa = 4.46LADD736 pKa = 3.86GSEE739 pKa = 4.29VKK741 pKa = 10.59KK742 pKa = 10.93GWIQIIRR749 pKa = 3.64
Molecular weight: 79.92 kDa
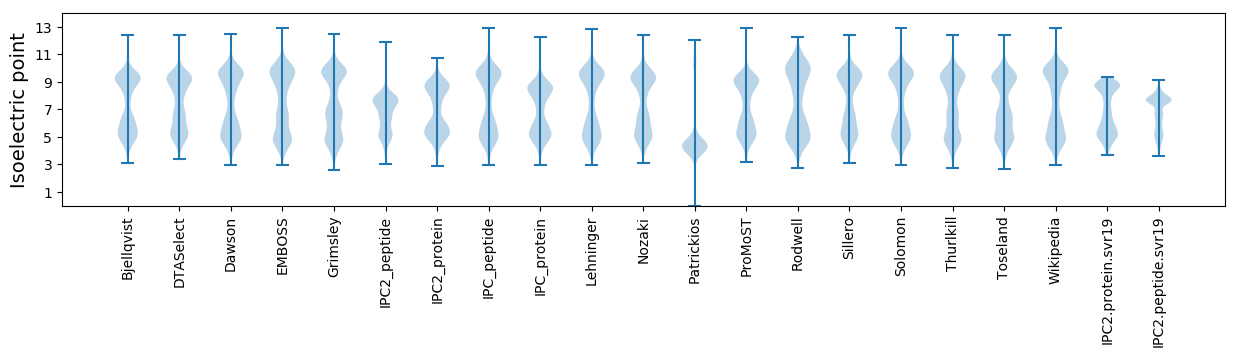
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0RGR8|F0RGR8_CELLC 50S ribosomal protein L32 OS=Cellulophaga lytica (strain ATCC 23178 / DSM 7489 / JCM 8516 / NBRC 14961 / NCIMB 1423 / VKM B-1433 / Cy l20) OX=867900 GN=rpmF PE=3 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.03GFRR20 pKa = 11.84EE21 pKa = 4.05RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.08GRR40 pKa = 11.84KK41 pKa = 8.3KK42 pKa = 9.93LTVSSEE48 pKa = 3.9LRR50 pKa = 11.84HH51 pKa = 5.93KK52 pKa = 10.53KK53 pKa = 10.04
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.03GFRR20 pKa = 11.84EE21 pKa = 4.05RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.08GRR40 pKa = 11.84KK41 pKa = 8.3KK42 pKa = 9.93LTVSSEE48 pKa = 3.9LRR50 pKa = 11.84HH51 pKa = 5.93KK52 pKa = 10.53KK53 pKa = 10.04
Molecular weight: 6.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1137891 |
31 |
4782 |
346.8 |
38.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.67 ± 0.048 | 0.717 ± 0.016 |
5.582 ± 0.054 | 6.311 ± 0.043 |
5.02 ± 0.034 | 6.315 ± 0.051 |
1.609 ± 0.019 | 7.691 ± 0.042 |
8.344 ± 0.077 | 9.202 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.016 ± 0.022 | 6.678 ± 0.044 |
3.286 ± 0.027 | 3.152 ± 0.024 |
3.022 ± 0.026 | 6.319 ± 0.034 |
6.421 ± 0.073 | 6.458 ± 0.036 |
1.014 ± 0.015 | 4.174 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
