
Clostridium ventriculi (Sarcina ventriculi)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firm
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
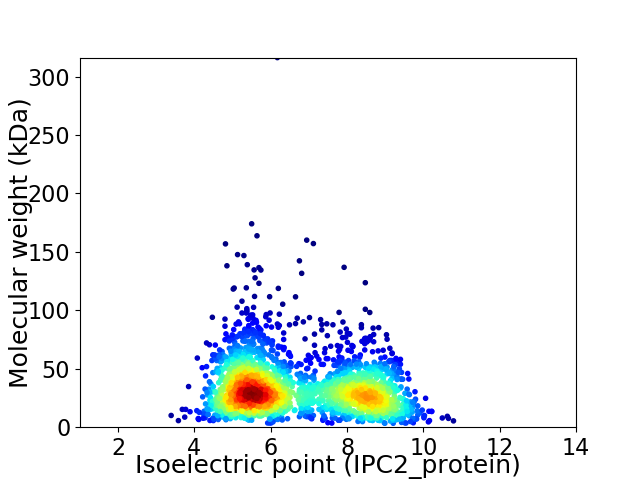
Virtual 2D-PAGE plot for 2228 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A174A8K5|A0A174A8K5_CLOVE Hypoxanthine phosphoribosyltransferase OS=Clostridium ventriculi OX=1267 GN=hpt_1 PE=3 SV=1
MM1 pKa = 7.53ACDD4 pKa = 3.36NTIQVLNGSVLTFEE18 pKa = 5.64SEE20 pKa = 4.27VQICNSITNLTKK32 pKa = 10.76LQFRR36 pKa = 11.84DD37 pKa = 3.29ILQGGLKK44 pKa = 8.78VTSDD48 pKa = 3.54QVNLTINNTSSTATAIVSEE67 pKa = 4.58TNNPEE72 pKa = 3.62DD73 pKa = 4.05TITVEE78 pKa = 4.1ILAANLGNDD87 pKa = 3.37YY88 pKa = 10.8PVTVKK93 pKa = 10.71VKK95 pKa = 10.06FQATVNDD102 pKa = 4.07LDD104 pKa = 4.57ALLEE108 pKa = 4.38NADD111 pKa = 4.37DD112 pKa = 3.82PTNAGKK118 pKa = 8.48LTNTGTFTALDD129 pKa = 3.63NAGASIGKK137 pKa = 8.8PATVTLADD145 pKa = 4.02TNVEE149 pKa = 4.22EE150 pKa = 4.49YY151 pKa = 10.59SPEE154 pKa = 3.71VSGYY158 pKa = 10.48SITVCPSEE166 pKa = 4.23NDD168 pKa = 3.31AKK170 pKa = 11.1FEE172 pKa = 4.94AKK174 pKa = 10.35LCLNTLCGDD183 pKa = 3.86YY184 pKa = 11.0TDD186 pKa = 4.11GKK188 pKa = 10.87YY189 pKa = 10.89VLDD192 pKa = 3.62VTAPDD197 pKa = 5.05GISFYY202 pKa = 11.21EE203 pKa = 4.75DD204 pKa = 3.01ATSTPPAAVEE214 pKa = 4.64SYY216 pKa = 9.76TGCCCKK222 pKa = 9.6STKK225 pKa = 9.74IEE227 pKa = 4.08MATPPVISADD237 pKa = 3.38NKK239 pKa = 9.65TVTITIASNEE249 pKa = 3.74VDD251 pKa = 3.4VALEE255 pKa = 3.87NDD257 pKa = 3.9TDD259 pKa = 4.24DD260 pKa = 3.86SCNNNISVYY269 pKa = 10.04IPCRR273 pKa = 11.84FDD275 pKa = 3.31NAASACTNEE284 pKa = 3.81PLIINGSIKK293 pKa = 10.07FTDD296 pKa = 3.72DD297 pKa = 3.08TDD299 pKa = 3.52NTLAEE304 pKa = 4.45LKK306 pKa = 11.03NFGLYY311 pKa = 10.92VNVDD315 pKa = 3.68CGNLVGVSKK324 pKa = 9.83TILCC328 pKa = 4.43
MM1 pKa = 7.53ACDD4 pKa = 3.36NTIQVLNGSVLTFEE18 pKa = 5.64SEE20 pKa = 4.27VQICNSITNLTKK32 pKa = 10.76LQFRR36 pKa = 11.84DD37 pKa = 3.29ILQGGLKK44 pKa = 8.78VTSDD48 pKa = 3.54QVNLTINNTSSTATAIVSEE67 pKa = 4.58TNNPEE72 pKa = 3.62DD73 pKa = 4.05TITVEE78 pKa = 4.1ILAANLGNDD87 pKa = 3.37YY88 pKa = 10.8PVTVKK93 pKa = 10.71VKK95 pKa = 10.06FQATVNDD102 pKa = 4.07LDD104 pKa = 4.57ALLEE108 pKa = 4.38NADD111 pKa = 4.37DD112 pKa = 3.82PTNAGKK118 pKa = 8.48LTNTGTFTALDD129 pKa = 3.63NAGASIGKK137 pKa = 8.8PATVTLADD145 pKa = 4.02TNVEE149 pKa = 4.22EE150 pKa = 4.49YY151 pKa = 10.59SPEE154 pKa = 3.71VSGYY158 pKa = 10.48SITVCPSEE166 pKa = 4.23NDD168 pKa = 3.31AKK170 pKa = 11.1FEE172 pKa = 4.94AKK174 pKa = 10.35LCLNTLCGDD183 pKa = 3.86YY184 pKa = 11.0TDD186 pKa = 4.11GKK188 pKa = 10.87YY189 pKa = 10.89VLDD192 pKa = 3.62VTAPDD197 pKa = 5.05GISFYY202 pKa = 11.21EE203 pKa = 4.75DD204 pKa = 3.01ATSTPPAAVEE214 pKa = 4.64SYY216 pKa = 9.76TGCCCKK222 pKa = 9.6STKK225 pKa = 9.74IEE227 pKa = 4.08MATPPVISADD237 pKa = 3.38NKK239 pKa = 9.65TVTITIASNEE249 pKa = 3.74VDD251 pKa = 3.4VALEE255 pKa = 3.87NDD257 pKa = 3.9TDD259 pKa = 4.24DD260 pKa = 3.86SCNNNISVYY269 pKa = 10.04IPCRR273 pKa = 11.84FDD275 pKa = 3.31NAASACTNEE284 pKa = 3.81PLIINGSIKK293 pKa = 10.07FTDD296 pKa = 3.72DD297 pKa = 3.08TDD299 pKa = 3.52NTLAEE304 pKa = 4.45LKK306 pKa = 11.03NFGLYY311 pKa = 10.92VNVDD315 pKa = 3.68CGNLVGVSKK324 pKa = 9.83TILCC328 pKa = 4.43
Molecular weight: 34.77 kDa
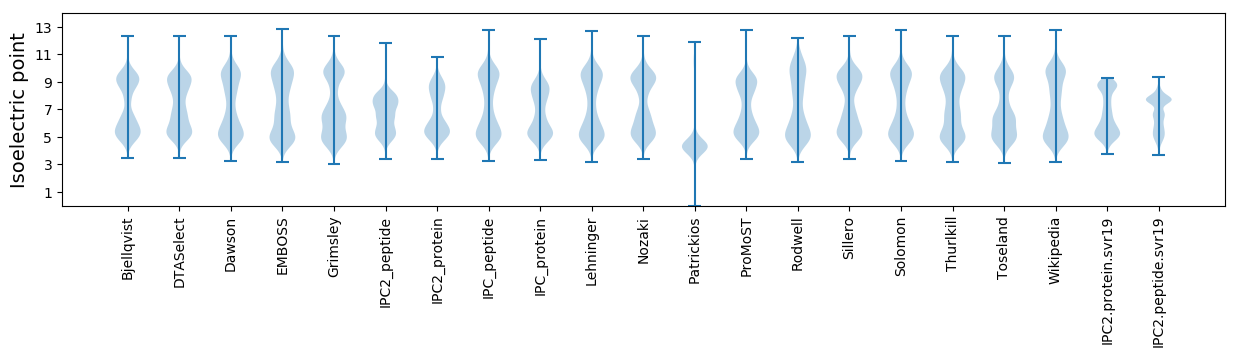
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A174BA25|A0A174BA25_CLOVE Oxygen-independent coproporphyrinogen-III oxidase 2 OS=Clostridium ventriculi OX=1267 GN=hemZ_2 PE=4 SV=1
MM1 pKa = 7.43SRR3 pKa = 11.84EE4 pKa = 3.27RR5 pKa = 11.84DD6 pKa = 3.31ARR8 pKa = 11.84KK9 pKa = 9.28GGKK12 pKa = 7.47MRR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84KK18 pKa = 8.94KK19 pKa = 9.33VCIFCVDD26 pKa = 3.15KK27 pKa = 11.51AEE29 pKa = 4.63SIDD32 pKa = 3.83YY33 pKa = 10.92KK34 pKa = 10.98DD35 pKa = 3.54VPKK38 pKa = 10.47LKK40 pKa = 10.42KK41 pKa = 10.69YY42 pKa = 7.85ITEE45 pKa = 4.07RR46 pKa = 11.84GKK48 pKa = 10.04ILPRR52 pKa = 11.84RR53 pKa = 11.84ISGTCAKK60 pKa = 9.97HH61 pKa = 5.4QRR63 pKa = 11.84QLTEE67 pKa = 4.3AIKK70 pKa = 10.27RR71 pKa = 11.84SRR73 pKa = 11.84NIALLPFTTEE83 pKa = 3.59
MM1 pKa = 7.43SRR3 pKa = 11.84EE4 pKa = 3.27RR5 pKa = 11.84DD6 pKa = 3.31ARR8 pKa = 11.84KK9 pKa = 9.28GGKK12 pKa = 7.47MRR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84KK18 pKa = 8.94KK19 pKa = 9.33VCIFCVDD26 pKa = 3.15KK27 pKa = 11.51AEE29 pKa = 4.63SIDD32 pKa = 3.83YY33 pKa = 10.92KK34 pKa = 10.98DD35 pKa = 3.54VPKK38 pKa = 10.47LKK40 pKa = 10.42KK41 pKa = 10.69YY42 pKa = 7.85ITEE45 pKa = 4.07RR46 pKa = 11.84GKK48 pKa = 10.04ILPRR52 pKa = 11.84RR53 pKa = 11.84ISGTCAKK60 pKa = 9.97HH61 pKa = 5.4QRR63 pKa = 11.84QLTEE67 pKa = 4.3AIKK70 pKa = 10.27RR71 pKa = 11.84SRR73 pKa = 11.84NIALLPFTTEE83 pKa = 3.59
Molecular weight: 9.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
702555 |
30 |
2730 |
315.3 |
35.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.361 ± 0.056 | 1.272 ± 0.021 |
5.512 ± 0.039 | 7.187 ± 0.069 |
4.589 ± 0.042 | 6.302 ± 0.051 |
1.289 ± 0.02 | 10.316 ± 0.064 |
9.311 ± 0.057 | 9.178 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.519 ± 0.026 | 6.961 ± 0.067 |
2.621 ± 0.028 | 2.15 ± 0.025 |
3.207 ± 0.037 | 6.262 ± 0.047 |
4.812 ± 0.045 | 6.414 ± 0.045 |
0.622 ± 0.016 | 4.113 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
