
Marinococcus luteus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Marinococcus
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
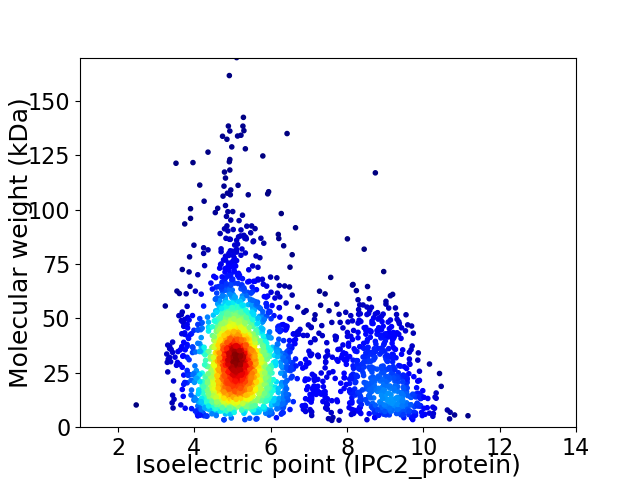
Virtual 2D-PAGE plot for 2897 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H2WA80|A0A1H2WA80_9BACI Two-component signal transduction system YycFG regulatory protein YycI OS=Marinococcus luteus OX=1122204 GN=SAMN05421781_2336 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 9.11EE3 pKa = 4.28TIKK6 pKa = 10.61SWTLSILVIGSLLLTWQNWSFQPEE30 pKa = 4.32YY31 pKa = 10.84EE32 pKa = 4.17NVNSSEE38 pKa = 4.34SVQEE42 pKa = 3.9TTEE45 pKa = 3.78IAEE48 pKa = 4.03TRR50 pKa = 11.84EE51 pKa = 3.94AGEE54 pKa = 4.29VVRR57 pKa = 11.84PTQGVVRR64 pKa = 11.84SNGSSSLMTEE74 pKa = 4.11DD75 pKa = 4.08QAASDD80 pKa = 4.0LDD82 pKa = 3.82KK83 pKa = 11.17VYY85 pKa = 10.57HH86 pKa = 5.57QLGEE90 pKa = 3.92ADD92 pKa = 3.17ITEE95 pKa = 4.32TSVTDD100 pKa = 3.95LDD102 pKa = 4.77HH103 pKa = 8.26DD104 pKa = 4.06EE105 pKa = 5.21LYY107 pKa = 11.19QNASSAEE114 pKa = 3.95YY115 pKa = 10.12SVEE118 pKa = 3.96LVFPSAVSWSMVNEE132 pKa = 4.18LFSFEE137 pKa = 4.85DD138 pKa = 3.55EE139 pKa = 4.45SNEE142 pKa = 4.19VEE144 pKa = 4.15EE145 pKa = 5.19ALDD148 pKa = 3.86GSIDD152 pKa = 4.32RR153 pKa = 11.84ILLTASPGSEE163 pKa = 3.69EE164 pKa = 3.99LNAVFVSYY172 pKa = 11.32DD173 pKa = 3.1EE174 pKa = 4.53GVVVNSNTNLDD185 pKa = 3.79KK186 pKa = 11.8NEE188 pKa = 4.06IEE190 pKa = 4.27YY191 pKa = 10.21EE192 pKa = 4.03QYY194 pKa = 10.83LAEE197 pKa = 4.61HH198 pKa = 6.92EE199 pKa = 4.58SMLTHH204 pKa = 7.0DD205 pKa = 4.65TDD207 pKa = 3.55QEE209 pKa = 4.66GYY211 pKa = 9.27EE212 pKa = 3.87KK213 pKa = 10.33HH214 pKa = 6.8IYY216 pKa = 9.92LPQDD220 pKa = 3.15PGARR224 pKa = 11.84TNYY227 pKa = 9.24TYY229 pKa = 11.51ASTDD233 pKa = 2.89LSTNEE238 pKa = 3.82FQQVLIADD246 pKa = 4.07EE247 pKa = 4.31NAQNLQTYY255 pKa = 8.43TSDD258 pKa = 3.26STEE261 pKa = 4.05VITDD265 pKa = 3.55GTRR268 pKa = 11.84EE269 pKa = 3.81LRR271 pKa = 11.84INNVGDD277 pKa = 3.47YY278 pKa = 10.81LRR280 pKa = 11.84YY281 pKa = 10.17ANPTYY286 pKa = 10.77SEE288 pKa = 4.11QTDD291 pKa = 3.96GNDD294 pKa = 3.25TSSLISSFEE303 pKa = 3.81FVNNHH308 pKa = 6.39AGWSDD313 pKa = 3.31EE314 pKa = 3.43YY315 pKa = 10.82RR316 pKa = 11.84YY317 pKa = 10.4YY318 pKa = 11.11SSSTVDD324 pKa = 4.02GEE326 pKa = 4.52TTDD329 pKa = 3.42TFRR332 pKa = 11.84MNKK335 pKa = 9.81GGLPVFRR342 pKa = 11.84DD343 pKa = 3.72SNGSDD348 pKa = 3.22FASIQVSNVGTQTTGYY364 pKa = 10.0SRR366 pKa = 11.84PLFDD370 pKa = 5.63LEE372 pKa = 4.29NNPVRR377 pKa = 11.84TGDD380 pKa = 3.58AGVSTDD386 pKa = 4.06LPTGEE391 pKa = 4.32EE392 pKa = 3.96TLDD395 pKa = 3.69AVQEE399 pKa = 4.25QCSSSLIEE407 pKa = 5.48DD408 pKa = 3.31IQIGYY413 pKa = 9.63SISRR417 pKa = 11.84NGEE420 pKa = 3.38NSGYY424 pKa = 10.74YY425 pKa = 10.47DD426 pKa = 4.17MEE428 pKa = 4.94PGWFVRR434 pKa = 11.84CDD436 pKa = 3.85GSWSPVEE443 pKa = 5.12FGGDD447 pKa = 3.49DD448 pKa = 3.62NEE450 pKa = 4.67EE451 pKa = 4.08GGEE454 pKa = 4.0
MM1 pKa = 7.68KK2 pKa = 9.11EE3 pKa = 4.28TIKK6 pKa = 10.61SWTLSILVIGSLLLTWQNWSFQPEE30 pKa = 4.32YY31 pKa = 10.84EE32 pKa = 4.17NVNSSEE38 pKa = 4.34SVQEE42 pKa = 3.9TTEE45 pKa = 3.78IAEE48 pKa = 4.03TRR50 pKa = 11.84EE51 pKa = 3.94AGEE54 pKa = 4.29VVRR57 pKa = 11.84PTQGVVRR64 pKa = 11.84SNGSSSLMTEE74 pKa = 4.11DD75 pKa = 4.08QAASDD80 pKa = 4.0LDD82 pKa = 3.82KK83 pKa = 11.17VYY85 pKa = 10.57HH86 pKa = 5.57QLGEE90 pKa = 3.92ADD92 pKa = 3.17ITEE95 pKa = 4.32TSVTDD100 pKa = 3.95LDD102 pKa = 4.77HH103 pKa = 8.26DD104 pKa = 4.06EE105 pKa = 5.21LYY107 pKa = 11.19QNASSAEE114 pKa = 3.95YY115 pKa = 10.12SVEE118 pKa = 3.96LVFPSAVSWSMVNEE132 pKa = 4.18LFSFEE137 pKa = 4.85DD138 pKa = 3.55EE139 pKa = 4.45SNEE142 pKa = 4.19VEE144 pKa = 4.15EE145 pKa = 5.19ALDD148 pKa = 3.86GSIDD152 pKa = 4.32RR153 pKa = 11.84ILLTASPGSEE163 pKa = 3.69EE164 pKa = 3.99LNAVFVSYY172 pKa = 11.32DD173 pKa = 3.1EE174 pKa = 4.53GVVVNSNTNLDD185 pKa = 3.79KK186 pKa = 11.8NEE188 pKa = 4.06IEE190 pKa = 4.27YY191 pKa = 10.21EE192 pKa = 4.03QYY194 pKa = 10.83LAEE197 pKa = 4.61HH198 pKa = 6.92EE199 pKa = 4.58SMLTHH204 pKa = 7.0DD205 pKa = 4.65TDD207 pKa = 3.55QEE209 pKa = 4.66GYY211 pKa = 9.27EE212 pKa = 3.87KK213 pKa = 10.33HH214 pKa = 6.8IYY216 pKa = 9.92LPQDD220 pKa = 3.15PGARR224 pKa = 11.84TNYY227 pKa = 9.24TYY229 pKa = 11.51ASTDD233 pKa = 2.89LSTNEE238 pKa = 3.82FQQVLIADD246 pKa = 4.07EE247 pKa = 4.31NAQNLQTYY255 pKa = 8.43TSDD258 pKa = 3.26STEE261 pKa = 4.05VITDD265 pKa = 3.55GTRR268 pKa = 11.84EE269 pKa = 3.81LRR271 pKa = 11.84INNVGDD277 pKa = 3.47YY278 pKa = 10.81LRR280 pKa = 11.84YY281 pKa = 10.17ANPTYY286 pKa = 10.77SEE288 pKa = 4.11QTDD291 pKa = 3.96GNDD294 pKa = 3.25TSSLISSFEE303 pKa = 3.81FVNNHH308 pKa = 6.39AGWSDD313 pKa = 3.31EE314 pKa = 3.43YY315 pKa = 10.82RR316 pKa = 11.84YY317 pKa = 10.4YY318 pKa = 11.11SSSTVDD324 pKa = 4.02GEE326 pKa = 4.52TTDD329 pKa = 3.42TFRR332 pKa = 11.84MNKK335 pKa = 9.81GGLPVFRR342 pKa = 11.84DD343 pKa = 3.72SNGSDD348 pKa = 3.22FASIQVSNVGTQTTGYY364 pKa = 10.0SRR366 pKa = 11.84PLFDD370 pKa = 5.63LEE372 pKa = 4.29NNPVRR377 pKa = 11.84TGDD380 pKa = 3.58AGVSTDD386 pKa = 4.06LPTGEE391 pKa = 4.32EE392 pKa = 3.96TLDD395 pKa = 3.69AVQEE399 pKa = 4.25QCSSSLIEE407 pKa = 5.48DD408 pKa = 3.31IQIGYY413 pKa = 9.63SISRR417 pKa = 11.84NGEE420 pKa = 3.38NSGYY424 pKa = 10.74YY425 pKa = 10.47DD426 pKa = 4.17MEE428 pKa = 4.94PGWFVRR434 pKa = 11.84CDD436 pKa = 3.85GSWSPVEE443 pKa = 5.12FGGDD447 pKa = 3.49DD448 pKa = 3.62NEE450 pKa = 4.67EE451 pKa = 4.08GGEE454 pKa = 4.0
Molecular weight: 50.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H2WHJ4|A0A1H2WHJ4_9BACI Glutamate synthase domain-containing protein 2 OS=Marinococcus luteus OX=1122204 GN=SAMN05421781_2414 PE=3 SV=1
MM1 pKa = 7.67GKK3 pKa = 7.99PTFTPNNRR11 pKa = 11.84KK12 pKa = 9.22RR13 pKa = 11.84KK14 pKa = 8.37KK15 pKa = 8.69VHH17 pKa = 5.54GFRR20 pKa = 11.84ARR22 pKa = 11.84MGTKK26 pKa = 10.01NGRR29 pKa = 11.84QVLKK33 pKa = 10.19NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.66GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
MM1 pKa = 7.67GKK3 pKa = 7.99PTFTPNNRR11 pKa = 11.84KK12 pKa = 9.22RR13 pKa = 11.84KK14 pKa = 8.37KK15 pKa = 8.69VHH17 pKa = 5.54GFRR20 pKa = 11.84ARR22 pKa = 11.84MGTKK26 pKa = 10.01NGRR29 pKa = 11.84QVLKK33 pKa = 10.19NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.66GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
842470 |
29 |
1535 |
290.8 |
32.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.352 ± 0.056 | 0.639 ± 0.012 |
5.211 ± 0.04 | 8.465 ± 0.061 |
4.229 ± 0.037 | 7.36 ± 0.044 |
2.26 ± 0.026 | 6.408 ± 0.046 |
5.256 ± 0.047 | 9.206 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.913 ± 0.022 | 3.719 ± 0.032 |
3.948 ± 0.023 | 3.953 ± 0.029 |
4.807 ± 0.037 | 6.034 ± 0.037 |
5.483 ± 0.028 | 7.198 ± 0.044 |
1.185 ± 0.018 | 3.374 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
