
Hubei tombus-like virus 34
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
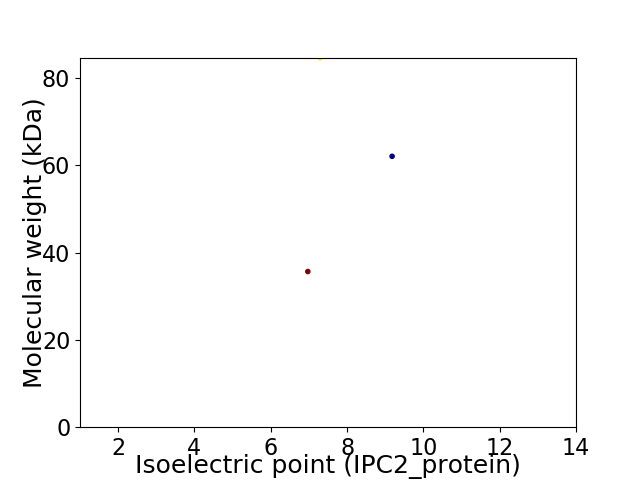
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KGT0|A0A1L3KGT0_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 34 OX=1923282 PE=4 SV=1
MM1 pKa = 7.24PTEE4 pKa = 3.88KK5 pKa = 10.65LYY7 pKa = 10.93DD8 pKa = 3.78EE9 pKa = 5.41RR10 pKa = 11.84DD11 pKa = 3.46GTNWDD16 pKa = 3.25STMQEE21 pKa = 3.22PHH23 pKa = 5.86MRR25 pKa = 11.84FEE27 pKa = 4.23ASVYY31 pKa = 10.74SLFDD35 pKa = 3.3AVIGQRR41 pKa = 11.84HH42 pKa = 5.5LQRR45 pKa = 11.84STHH48 pKa = 5.99CKK50 pKa = 8.48GTIATRR56 pKa = 11.84DD57 pKa = 3.7YY58 pKa = 10.32IIKK61 pKa = 10.55YY62 pKa = 4.66VTRR65 pKa = 11.84WKK67 pKa = 10.53RR68 pKa = 11.84LSGDD72 pKa = 2.86WNTSVGNSIISMAICITAILSLPSHH97 pKa = 6.62LRR99 pKa = 11.84PHH101 pKa = 6.36RR102 pKa = 11.84VAAFFHH108 pKa = 6.69GDD110 pKa = 3.45DD111 pKa = 3.64YY112 pKa = 11.78LAIYY116 pKa = 9.02HH117 pKa = 5.46YY118 pKa = 10.33HH119 pKa = 7.2RR120 pKa = 11.84SPPPQVLNTALADD133 pKa = 3.85LEE135 pKa = 4.32KK136 pKa = 10.93SLGITPVRR144 pKa = 11.84GVFRR148 pKa = 11.84DD149 pKa = 3.74PLLVEE154 pKa = 4.51YY155 pKa = 10.24ISMSVWPCYY164 pKa = 10.44DD165 pKa = 2.74GTYY168 pKa = 10.53FFAPKK173 pKa = 10.3LSNLFVRR180 pKa = 11.84LFFSTRR186 pKa = 11.84PLSQHH191 pKa = 5.74TADD194 pKa = 4.92DD195 pKa = 3.85VCATIAALAPHH206 pKa = 6.58FVGCEE211 pKa = 3.33PAEE214 pKa = 4.1RR215 pKa = 11.84FFAAHH220 pKa = 5.51RR221 pKa = 11.84RR222 pKa = 11.84AWIARR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84GIKK232 pKa = 9.78HH233 pKa = 6.57AGNQFRR239 pKa = 11.84DD240 pKa = 3.73CHH242 pKa = 7.07SLEE245 pKa = 4.2KK246 pKa = 11.09LEE248 pKa = 4.77IDD250 pKa = 3.81HH251 pKa = 6.41TPKK254 pKa = 10.41VHH256 pKa = 5.92WAYY259 pKa = 10.7GFAHH263 pKa = 7.76KK264 pKa = 10.92YY265 pKa = 9.82HH266 pKa = 6.15MPITALHH273 pKa = 6.0VEE275 pKa = 4.56VEE277 pKa = 4.45CGSAALLRR285 pKa = 11.84HH286 pKa = 6.2PAYY289 pKa = 10.54DD290 pKa = 3.35HH291 pKa = 7.1LFEE294 pKa = 6.05CEE296 pKa = 4.29HH297 pKa = 7.09LDD299 pKa = 3.31PDD301 pKa = 3.8VRR303 pKa = 11.84MGASATRR310 pKa = 11.84DD311 pKa = 2.97II312 pKa = 4.9
MM1 pKa = 7.24PTEE4 pKa = 3.88KK5 pKa = 10.65LYY7 pKa = 10.93DD8 pKa = 3.78EE9 pKa = 5.41RR10 pKa = 11.84DD11 pKa = 3.46GTNWDD16 pKa = 3.25STMQEE21 pKa = 3.22PHH23 pKa = 5.86MRR25 pKa = 11.84FEE27 pKa = 4.23ASVYY31 pKa = 10.74SLFDD35 pKa = 3.3AVIGQRR41 pKa = 11.84HH42 pKa = 5.5LQRR45 pKa = 11.84STHH48 pKa = 5.99CKK50 pKa = 8.48GTIATRR56 pKa = 11.84DD57 pKa = 3.7YY58 pKa = 10.32IIKK61 pKa = 10.55YY62 pKa = 4.66VTRR65 pKa = 11.84WKK67 pKa = 10.53RR68 pKa = 11.84LSGDD72 pKa = 2.86WNTSVGNSIISMAICITAILSLPSHH97 pKa = 6.62LRR99 pKa = 11.84PHH101 pKa = 6.36RR102 pKa = 11.84VAAFFHH108 pKa = 6.69GDD110 pKa = 3.45DD111 pKa = 3.64YY112 pKa = 11.78LAIYY116 pKa = 9.02HH117 pKa = 5.46YY118 pKa = 10.33HH119 pKa = 7.2RR120 pKa = 11.84SPPPQVLNTALADD133 pKa = 3.85LEE135 pKa = 4.32KK136 pKa = 10.93SLGITPVRR144 pKa = 11.84GVFRR148 pKa = 11.84DD149 pKa = 3.74PLLVEE154 pKa = 4.51YY155 pKa = 10.24ISMSVWPCYY164 pKa = 10.44DD165 pKa = 2.74GTYY168 pKa = 10.53FFAPKK173 pKa = 10.3LSNLFVRR180 pKa = 11.84LFFSTRR186 pKa = 11.84PLSQHH191 pKa = 5.74TADD194 pKa = 4.92DD195 pKa = 3.85VCATIAALAPHH206 pKa = 6.58FVGCEE211 pKa = 3.33PAEE214 pKa = 4.1RR215 pKa = 11.84FFAAHH220 pKa = 5.51RR221 pKa = 11.84RR222 pKa = 11.84AWIARR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84GIKK232 pKa = 9.78HH233 pKa = 6.57AGNQFRR239 pKa = 11.84DD240 pKa = 3.73CHH242 pKa = 7.07SLEE245 pKa = 4.2KK246 pKa = 11.09LEE248 pKa = 4.77IDD250 pKa = 3.81HH251 pKa = 6.41TPKK254 pKa = 10.41VHH256 pKa = 5.92WAYY259 pKa = 10.7GFAHH263 pKa = 7.76KK264 pKa = 10.92YY265 pKa = 9.82HH266 pKa = 6.15MPITALHH273 pKa = 6.0VEE275 pKa = 4.56VEE277 pKa = 4.45CGSAALLRR285 pKa = 11.84HH286 pKa = 6.2PAYY289 pKa = 10.54DD290 pKa = 3.35HH291 pKa = 7.1LFEE294 pKa = 6.05CEE296 pKa = 4.29HH297 pKa = 7.09LDD299 pKa = 3.31PDD301 pKa = 3.8VRR303 pKa = 11.84MGASATRR310 pKa = 11.84DD311 pKa = 2.97II312 pKa = 4.9
Molecular weight: 35.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGM2|A0A1L3KGM2_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 34 OX=1923282 PE=4 SV=1
MM1 pKa = 7.49LEE3 pKa = 3.46WGRR6 pKa = 11.84PRR8 pKa = 11.84RR9 pKa = 11.84EE10 pKa = 3.89TSDD13 pKa = 2.93SDD15 pKa = 3.44LRR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.82IEE21 pKa = 3.99TMATNLARR29 pKa = 11.84AIALPGKK36 pKa = 9.19HH37 pKa = 6.08APLRR41 pKa = 11.84FPSFPALEE49 pKa = 4.21RR50 pKa = 11.84TAVMAFNASMPFSIGADD67 pKa = 3.39STKK70 pKa = 10.79VILTRR75 pKa = 11.84QAALPLWGEE84 pKa = 4.21TKK86 pKa = 10.89APGGEE91 pKa = 4.1NAALTYY97 pKa = 10.48GVVYY101 pKa = 10.64SCTGVSAVNTQGGIYY116 pKa = 9.45TEE118 pKa = 4.31AVDD121 pKa = 6.29AIGGWHH127 pKa = 5.83QSVTYY132 pKa = 10.11GLSAPVMAVPGGTWPTSYY150 pKa = 10.53PVVAFDD156 pKa = 4.98AATGDD161 pKa = 4.35LPWVYY166 pKa = 10.1CPNGGKK172 pKa = 9.62VLITSNAMAFAHH184 pKa = 6.63NDD186 pKa = 3.44NIQLEE191 pKa = 4.67RR192 pKa = 11.84WTGPGQSEE200 pKa = 5.22PITLTAAHH208 pKa = 6.25TASLNLGGGSYY219 pKa = 11.04FDD221 pKa = 3.39VTANAWYY228 pKa = 10.0RR229 pKa = 11.84FRR231 pKa = 11.84SSSIQDD237 pKa = 3.36TTTGSVLKK245 pKa = 7.51FTPVYY250 pKa = 10.44SLGVLLSDD258 pKa = 4.13QAPTFTGAAVNGTWAIPTTNDD279 pKa = 2.56QRR281 pKa = 11.84RR282 pKa = 11.84FLLPLTVSPEE292 pKa = 3.94FSNSHH297 pKa = 6.19LPWAATRR304 pKa = 11.84LTAVGALFTNTTKK317 pKa = 10.7VLNKK321 pKa = 9.95EE322 pKa = 4.28GTVLWGRR329 pKa = 11.84INPVVYY335 pKa = 10.48DD336 pKa = 3.83PFIVTSTTIQNLHH349 pKa = 6.37PAEE352 pKa = 4.1KK353 pKa = 10.16AYY355 pKa = 11.1LDD357 pKa = 4.54LEE359 pKa = 4.95HH360 pKa = 6.68GTYY363 pKa = 10.29AYY365 pKa = 10.38NPPSTDD371 pKa = 3.25LADD374 pKa = 3.83FTPYY378 pKa = 10.36ILRR381 pKa = 11.84LLPNSASNSAIWPVYY396 pKa = 10.28RR397 pKa = 11.84LDD399 pKa = 3.48NRR401 pKa = 11.84AFAAVGFFNDD411 pKa = 3.99PDD413 pKa = 4.28GGTNLAVNLDD423 pKa = 3.46YY424 pKa = 10.97HH425 pKa = 7.98IEE427 pKa = 4.02FRR429 pKa = 11.84TSSTLFQIGVSTMPLEE445 pKa = 4.5ALHH448 pKa = 6.49AAQIVLLKK456 pKa = 10.92AGFFFHH462 pKa = 7.08NEE464 pKa = 3.4DD465 pKa = 3.36HH466 pKa = 6.55GAVIARR472 pKa = 11.84IVKK475 pKa = 9.39FMAGMHH481 pKa = 6.92PLLSMAVPIANGLIGASSYY500 pKa = 11.6ALSSKK505 pKa = 9.96PNQSHH510 pKa = 5.78TPAATSGQGAGMVSVAAPPRR530 pKa = 11.84PRR532 pKa = 11.84RR533 pKa = 11.84AAARR537 pKa = 11.84RR538 pKa = 11.84PPARR542 pKa = 11.84ARR544 pKa = 11.84PRR546 pKa = 11.84PRR548 pKa = 11.84RR549 pKa = 11.84AAPPPPPPPQKK560 pKa = 10.62KK561 pKa = 9.6KK562 pKa = 10.91GKK564 pKa = 9.06LASGLDD570 pKa = 3.58MYY572 pKa = 11.16LSRR575 pKa = 11.84KK576 pKa = 8.94KK577 pKa = 10.5RR578 pKa = 3.36
MM1 pKa = 7.49LEE3 pKa = 3.46WGRR6 pKa = 11.84PRR8 pKa = 11.84RR9 pKa = 11.84EE10 pKa = 3.89TSDD13 pKa = 2.93SDD15 pKa = 3.44LRR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.82IEE21 pKa = 3.99TMATNLARR29 pKa = 11.84AIALPGKK36 pKa = 9.19HH37 pKa = 6.08APLRR41 pKa = 11.84FPSFPALEE49 pKa = 4.21RR50 pKa = 11.84TAVMAFNASMPFSIGADD67 pKa = 3.39STKK70 pKa = 10.79VILTRR75 pKa = 11.84QAALPLWGEE84 pKa = 4.21TKK86 pKa = 10.89APGGEE91 pKa = 4.1NAALTYY97 pKa = 10.48GVVYY101 pKa = 10.64SCTGVSAVNTQGGIYY116 pKa = 9.45TEE118 pKa = 4.31AVDD121 pKa = 6.29AIGGWHH127 pKa = 5.83QSVTYY132 pKa = 10.11GLSAPVMAVPGGTWPTSYY150 pKa = 10.53PVVAFDD156 pKa = 4.98AATGDD161 pKa = 4.35LPWVYY166 pKa = 10.1CPNGGKK172 pKa = 9.62VLITSNAMAFAHH184 pKa = 6.63NDD186 pKa = 3.44NIQLEE191 pKa = 4.67RR192 pKa = 11.84WTGPGQSEE200 pKa = 5.22PITLTAAHH208 pKa = 6.25TASLNLGGGSYY219 pKa = 11.04FDD221 pKa = 3.39VTANAWYY228 pKa = 10.0RR229 pKa = 11.84FRR231 pKa = 11.84SSSIQDD237 pKa = 3.36TTTGSVLKK245 pKa = 7.51FTPVYY250 pKa = 10.44SLGVLLSDD258 pKa = 4.13QAPTFTGAAVNGTWAIPTTNDD279 pKa = 2.56QRR281 pKa = 11.84RR282 pKa = 11.84FLLPLTVSPEE292 pKa = 3.94FSNSHH297 pKa = 6.19LPWAATRR304 pKa = 11.84LTAVGALFTNTTKK317 pKa = 10.7VLNKK321 pKa = 9.95EE322 pKa = 4.28GTVLWGRR329 pKa = 11.84INPVVYY335 pKa = 10.48DD336 pKa = 3.83PFIVTSTTIQNLHH349 pKa = 6.37PAEE352 pKa = 4.1KK353 pKa = 10.16AYY355 pKa = 11.1LDD357 pKa = 4.54LEE359 pKa = 4.95HH360 pKa = 6.68GTYY363 pKa = 10.29AYY365 pKa = 10.38NPPSTDD371 pKa = 3.25LADD374 pKa = 3.83FTPYY378 pKa = 10.36ILRR381 pKa = 11.84LLPNSASNSAIWPVYY396 pKa = 10.28RR397 pKa = 11.84LDD399 pKa = 3.48NRR401 pKa = 11.84AFAAVGFFNDD411 pKa = 3.99PDD413 pKa = 4.28GGTNLAVNLDD423 pKa = 3.46YY424 pKa = 10.97HH425 pKa = 7.98IEE427 pKa = 4.02FRR429 pKa = 11.84TSSTLFQIGVSTMPLEE445 pKa = 4.5ALHH448 pKa = 6.49AAQIVLLKK456 pKa = 10.92AGFFFHH462 pKa = 7.08NEE464 pKa = 3.4DD465 pKa = 3.36HH466 pKa = 6.55GAVIARR472 pKa = 11.84IVKK475 pKa = 9.39FMAGMHH481 pKa = 6.92PLLSMAVPIANGLIGASSYY500 pKa = 11.6ALSSKK505 pKa = 9.96PNQSHH510 pKa = 5.78TPAATSGQGAGMVSVAAPPRR530 pKa = 11.84PRR532 pKa = 11.84RR533 pKa = 11.84AAARR537 pKa = 11.84RR538 pKa = 11.84PPARR542 pKa = 11.84ARR544 pKa = 11.84PRR546 pKa = 11.84PRR548 pKa = 11.84RR549 pKa = 11.84AAPPPPPPPQKK560 pKa = 10.62KK561 pKa = 9.6KK562 pKa = 10.91GKK564 pKa = 9.06LASGLDD570 pKa = 3.58MYY572 pKa = 11.16LSRR575 pKa = 11.84KK576 pKa = 8.94KK577 pKa = 10.5RR578 pKa = 3.36
Molecular weight: 62.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1665 |
312 |
775 |
555.0 |
60.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.793 ± 0.973 | 1.502 ± 0.484 |
4.865 ± 0.52 | 4.144 ± 0.607 |
3.844 ± 0.464 | 6.426 ± 0.592 |
3.183 ± 0.924 | 3.844 ± 0.599 |
3.483 ± 0.224 | 8.228 ± 0.296 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.462 ± 0.219 | 3.724 ± 0.509 |
6.486 ± 0.938 | 2.763 ± 0.352 |
6.667 ± 0.409 | 6.066 ± 0.619 |
7.147 ± 0.565 | 7.327 ± 0.917 |
1.802 ± 0.072 | 3.243 ± 0.249 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
