
Sphingomonas sp. EC-HK361
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
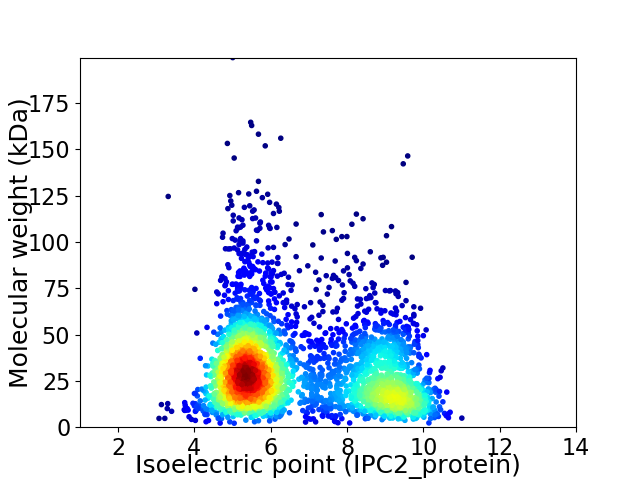
Virtual 2D-PAGE plot for 3256 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5E7XR96|A0A5E7XR96_9SPHN 50S ribosomal protein L33 OS=Sphingomonas sp. EC-HK361 OX=2038397 GN=rpmG PE=3 SV=1
MM1 pKa = 7.69ADD3 pKa = 3.42YY4 pKa = 11.22YY5 pKa = 10.36NTLGSSIANGTTGRR19 pKa = 11.84DD20 pKa = 3.11LFFPFTKK27 pKa = 10.48DD28 pKa = 3.07PNFDD32 pKa = 3.59HH33 pKa = 6.92LRR35 pKa = 11.84ADD37 pKa = 3.39ATLSQLVWSTGILTGSGTAYY57 pKa = 10.43QIIAPNVQISMDD69 pKa = 4.08LLNGGEE75 pKa = 4.37GTDD78 pKa = 3.04IVYY81 pKa = 10.85ASNVADD87 pKa = 3.59ALFYY91 pKa = 11.42NNGAIAGGFGGFSNIEE107 pKa = 4.05QFWMGDD113 pKa = 2.94GDD115 pKa = 5.82DD116 pKa = 5.21IIDD119 pKa = 4.09LTAHH123 pKa = 6.61GAGGVDD129 pKa = 3.65YY130 pKa = 10.87AKK132 pKa = 10.63AAAIHH137 pKa = 5.57GQGGNDD143 pKa = 3.75IIIGGAGKK151 pKa = 8.58DD152 pKa = 3.66TLEE155 pKa = 4.73GDD157 pKa = 3.7AGNDD161 pKa = 3.57IIFGWRR167 pKa = 11.84GSDD170 pKa = 3.44TIFGGTGDD178 pKa = 4.37DD179 pKa = 4.74LLYY182 pKa = 11.19GDD184 pKa = 5.62DD185 pKa = 4.73LGFNGIAGDD194 pKa = 3.82DD195 pKa = 4.25TIDD198 pKa = 3.63GGAGNDD204 pKa = 3.05ILYY207 pKa = 10.5GGRR210 pKa = 11.84GSDD213 pKa = 4.0KK214 pKa = 9.3MTGGDD219 pKa = 4.91DD220 pKa = 3.81NDD222 pKa = 3.64ILYY225 pKa = 9.93GQAGGDD231 pKa = 3.61NMSGGSGDD239 pKa = 4.01DD240 pKa = 3.87LLYY243 pKa = 11.2GDD245 pKa = 5.82DD246 pKa = 6.16ADD248 pKa = 4.14TTSNDD253 pKa = 3.82TLNGDD258 pKa = 3.66AGADD262 pKa = 3.38RR263 pKa = 11.84LYY265 pKa = 11.28GGGGDD270 pKa = 3.86DD271 pKa = 3.82EE272 pKa = 6.14LYY274 pKa = 10.98GGSGDD279 pKa = 4.87DD280 pKa = 3.99YY281 pKa = 11.7LDD283 pKa = 4.0GGTGNDD289 pKa = 3.88YY290 pKa = 11.25VHH292 pKa = 7.03GGAGNDD298 pKa = 3.89TIVAGAGNDD307 pKa = 3.87IIDD310 pKa = 4.19GSADD314 pKa = 2.97IDD316 pKa = 3.79TVVFAGNRR324 pKa = 11.84ADD326 pKa = 3.69YY327 pKa = 10.86QFALQTDD334 pKa = 4.46GSFIATDD341 pKa = 3.44MRR343 pKa = 11.84GGSPEE348 pKa = 3.8GTKK351 pKa = 9.94TIRR354 pKa = 11.84NVEE357 pKa = 4.28FFAFADD363 pKa = 3.94TTVSSTLLNYY373 pKa = 10.67LPVITSNGSGAAASLDD389 pKa = 3.17IDD391 pKa = 4.05EE392 pKa = 5.05NATLVTTVTATDD404 pKa = 3.15QDD406 pKa = 3.52AGQTLTFTIAGGADD420 pKa = 2.98AALFTVDD427 pKa = 3.99PVTGVLSFITAPNFEE442 pKa = 4.79SPSDD446 pKa = 3.79ANGDD450 pKa = 3.2NVYY453 pKa = 10.48RR454 pKa = 11.84VVVAVNDD461 pKa = 4.03GNGGVDD467 pKa = 3.55TQDD470 pKa = 3.26LSVNVRR476 pKa = 11.84DD477 pKa = 4.31VPDD480 pKa = 3.8GFAPVITSNGGGATAAITVNEE501 pKa = 4.14NLTAVTTVTATDD513 pKa = 3.57ADD515 pKa = 4.32GPTLAFAIIGGADD528 pKa = 3.18AALFAIDD535 pKa = 4.43PATGALTFKK544 pKa = 10.41NAPDD548 pKa = 3.77FEE550 pKa = 4.76NPGDD554 pKa = 3.98ANHH557 pKa = 7.2DD558 pKa = 3.51NVYY561 pKa = 10.92DD562 pKa = 3.98VVVQASDD569 pKa = 3.62GNNTDD574 pKa = 3.26SQALSVTVGNVNDD587 pKa = 3.95NAPALTSFGGALAASFTVNEE607 pKa = 4.36NVTAATTIAASDD619 pKa = 3.78ADD621 pKa = 4.33GDD623 pKa = 4.21ALSYY627 pKa = 10.43TITGGADD634 pKa = 2.9AALFAVDD641 pKa = 3.78AATGRR646 pKa = 11.84LTFITPPDD654 pKa = 3.61YY655 pKa = 10.2EE656 pKa = 5.0APGDD660 pKa = 3.87SDD662 pKa = 3.62HH663 pKa = 7.38DD664 pKa = 4.13RR665 pKa = 11.84IYY667 pKa = 10.93QVVVSASDD675 pKa = 3.47GTNSVAQALSITIANVNDD693 pKa = 3.65NAPVITSNGGGGTASLGIAEE713 pKa = 5.14NGTAVTIVTATDD725 pKa = 3.39ADD727 pKa = 4.23GTVPTYY733 pKa = 10.87AIAGGADD740 pKa = 3.06AALFTIDD747 pKa = 4.22PATGALSFLSAPDD760 pKa = 3.82FEE762 pKa = 6.09HH763 pKa = 7.51PLDD766 pKa = 4.46ADD768 pKa = 3.65ANGIYY773 pKa = 9.89QVTVRR778 pKa = 11.84ATDD781 pKa = 3.44GLNVVDD787 pKa = 4.41QLLSVSVTDD796 pKa = 3.47VNEE799 pKa = 3.91VGRR802 pKa = 11.84TITGTSGNNTISPTTTVVGYY822 pKa = 7.47QTTALGDD829 pKa = 3.7TIYY832 pKa = 11.41ALAGNDD838 pKa = 4.05IIDD841 pKa = 4.14GGAGADD847 pKa = 3.9YY848 pKa = 10.48MDD850 pKa = 4.95GGAGNDD856 pKa = 3.35TFYY859 pKa = 11.74VDD861 pKa = 3.46TFSDD865 pKa = 3.66DD866 pKa = 3.9GYY868 pKa = 11.22AGNDD872 pKa = 3.32DD873 pKa = 4.49RR874 pKa = 11.84VIEE877 pKa = 4.03LAGGGTDD884 pKa = 3.68LVYY887 pKa = 11.06ASVSYY892 pKa = 10.8RR893 pKa = 11.84LADD896 pKa = 3.39QVEE899 pKa = 4.18NLTLTGTAALNGYY912 pKa = 10.21GNDD915 pKa = 3.54LANIITGNDD924 pKa = 2.96AANRR928 pKa = 11.84LEE930 pKa = 4.55GGLGADD936 pKa = 3.68TLYY939 pKa = 11.55GMGGNDD945 pKa = 3.7TLDD948 pKa = 3.8GGDD951 pKa = 3.91GNDD954 pKa = 3.41RR955 pKa = 11.84LYY957 pKa = 11.36GGDD960 pKa = 3.73GTDD963 pKa = 3.38TLIGGTGDD971 pKa = 3.49DD972 pKa = 4.27TLDD975 pKa = 3.78GGTGADD981 pKa = 3.6TMTGGDD987 pKa = 4.22GNDD990 pKa = 3.1SYY992 pKa = 12.01YY993 pKa = 11.05VDD995 pKa = 4.2DD996 pKa = 5.85AGDD999 pKa = 3.63QVVEE1003 pKa = 4.18LANGGTTDD1011 pKa = 3.99QIFSAITYY1019 pKa = 7.53SLPAEE1024 pKa = 4.3VEE1026 pKa = 4.14KK1027 pKa = 10.44LTLTGTDD1034 pKa = 3.89AIDD1037 pKa = 3.79GRR1039 pKa = 11.84GNEE1042 pKa = 4.47LNNTITGNGANNGLWGGAGNDD1063 pKa = 3.89TLSGNAGDD1071 pKa = 4.98DD1072 pKa = 3.5RR1073 pKa = 11.84LFGEE1077 pKa = 5.45DD1078 pKa = 5.3GLDD1081 pKa = 3.53TLDD1084 pKa = 4.18GGVGNDD1090 pKa = 3.94RR1091 pKa = 11.84LDD1093 pKa = 3.88GGAGADD1099 pKa = 3.68TLLGKK1104 pKa = 10.31AGNDD1108 pKa = 3.61TLIGGAGKK1116 pKa = 8.54DD1117 pKa = 3.47TLTGGTEE1124 pKa = 3.76ADD1126 pKa = 3.14TFVFNFGDD1134 pKa = 3.64TTLNSASYY1142 pKa = 11.18DD1143 pKa = 3.65RR1144 pKa = 11.84ITDD1147 pKa = 4.24FKK1149 pKa = 11.21ASEE1152 pKa = 4.3GDD1154 pKa = 4.06VIDD1157 pKa = 5.0LDD1159 pKa = 4.22VVNGSLAPADD1169 pKa = 4.04FTATTIATNNFADD1182 pKa = 4.09ALTAAKK1188 pKa = 8.54LTQSAGHH1195 pKa = 5.64ITFVAGATDD1204 pKa = 4.41GWLFWDD1210 pKa = 4.38GNGDD1214 pKa = 3.83GVFDD1218 pKa = 4.41QSVLLSGANTQSAIDD1233 pKa = 3.94PLHH1236 pKa = 6.27IVV1238 pKa = 3.39
MM1 pKa = 7.69ADD3 pKa = 3.42YY4 pKa = 11.22YY5 pKa = 10.36NTLGSSIANGTTGRR19 pKa = 11.84DD20 pKa = 3.11LFFPFTKK27 pKa = 10.48DD28 pKa = 3.07PNFDD32 pKa = 3.59HH33 pKa = 6.92LRR35 pKa = 11.84ADD37 pKa = 3.39ATLSQLVWSTGILTGSGTAYY57 pKa = 10.43QIIAPNVQISMDD69 pKa = 4.08LLNGGEE75 pKa = 4.37GTDD78 pKa = 3.04IVYY81 pKa = 10.85ASNVADD87 pKa = 3.59ALFYY91 pKa = 11.42NNGAIAGGFGGFSNIEE107 pKa = 4.05QFWMGDD113 pKa = 2.94GDD115 pKa = 5.82DD116 pKa = 5.21IIDD119 pKa = 4.09LTAHH123 pKa = 6.61GAGGVDD129 pKa = 3.65YY130 pKa = 10.87AKK132 pKa = 10.63AAAIHH137 pKa = 5.57GQGGNDD143 pKa = 3.75IIIGGAGKK151 pKa = 8.58DD152 pKa = 3.66TLEE155 pKa = 4.73GDD157 pKa = 3.7AGNDD161 pKa = 3.57IIFGWRR167 pKa = 11.84GSDD170 pKa = 3.44TIFGGTGDD178 pKa = 4.37DD179 pKa = 4.74LLYY182 pKa = 11.19GDD184 pKa = 5.62DD185 pKa = 4.73LGFNGIAGDD194 pKa = 3.82DD195 pKa = 4.25TIDD198 pKa = 3.63GGAGNDD204 pKa = 3.05ILYY207 pKa = 10.5GGRR210 pKa = 11.84GSDD213 pKa = 4.0KK214 pKa = 9.3MTGGDD219 pKa = 4.91DD220 pKa = 3.81NDD222 pKa = 3.64ILYY225 pKa = 9.93GQAGGDD231 pKa = 3.61NMSGGSGDD239 pKa = 4.01DD240 pKa = 3.87LLYY243 pKa = 11.2GDD245 pKa = 5.82DD246 pKa = 6.16ADD248 pKa = 4.14TTSNDD253 pKa = 3.82TLNGDD258 pKa = 3.66AGADD262 pKa = 3.38RR263 pKa = 11.84LYY265 pKa = 11.28GGGGDD270 pKa = 3.86DD271 pKa = 3.82EE272 pKa = 6.14LYY274 pKa = 10.98GGSGDD279 pKa = 4.87DD280 pKa = 3.99YY281 pKa = 11.7LDD283 pKa = 4.0GGTGNDD289 pKa = 3.88YY290 pKa = 11.25VHH292 pKa = 7.03GGAGNDD298 pKa = 3.89TIVAGAGNDD307 pKa = 3.87IIDD310 pKa = 4.19GSADD314 pKa = 2.97IDD316 pKa = 3.79TVVFAGNRR324 pKa = 11.84ADD326 pKa = 3.69YY327 pKa = 10.86QFALQTDD334 pKa = 4.46GSFIATDD341 pKa = 3.44MRR343 pKa = 11.84GGSPEE348 pKa = 3.8GTKK351 pKa = 9.94TIRR354 pKa = 11.84NVEE357 pKa = 4.28FFAFADD363 pKa = 3.94TTVSSTLLNYY373 pKa = 10.67LPVITSNGSGAAASLDD389 pKa = 3.17IDD391 pKa = 4.05EE392 pKa = 5.05NATLVTTVTATDD404 pKa = 3.15QDD406 pKa = 3.52AGQTLTFTIAGGADD420 pKa = 2.98AALFTVDD427 pKa = 3.99PVTGVLSFITAPNFEE442 pKa = 4.79SPSDD446 pKa = 3.79ANGDD450 pKa = 3.2NVYY453 pKa = 10.48RR454 pKa = 11.84VVVAVNDD461 pKa = 4.03GNGGVDD467 pKa = 3.55TQDD470 pKa = 3.26LSVNVRR476 pKa = 11.84DD477 pKa = 4.31VPDD480 pKa = 3.8GFAPVITSNGGGATAAITVNEE501 pKa = 4.14NLTAVTTVTATDD513 pKa = 3.57ADD515 pKa = 4.32GPTLAFAIIGGADD528 pKa = 3.18AALFAIDD535 pKa = 4.43PATGALTFKK544 pKa = 10.41NAPDD548 pKa = 3.77FEE550 pKa = 4.76NPGDD554 pKa = 3.98ANHH557 pKa = 7.2DD558 pKa = 3.51NVYY561 pKa = 10.92DD562 pKa = 3.98VVVQASDD569 pKa = 3.62GNNTDD574 pKa = 3.26SQALSVTVGNVNDD587 pKa = 3.95NAPALTSFGGALAASFTVNEE607 pKa = 4.36NVTAATTIAASDD619 pKa = 3.78ADD621 pKa = 4.33GDD623 pKa = 4.21ALSYY627 pKa = 10.43TITGGADD634 pKa = 2.9AALFAVDD641 pKa = 3.78AATGRR646 pKa = 11.84LTFITPPDD654 pKa = 3.61YY655 pKa = 10.2EE656 pKa = 5.0APGDD660 pKa = 3.87SDD662 pKa = 3.62HH663 pKa = 7.38DD664 pKa = 4.13RR665 pKa = 11.84IYY667 pKa = 10.93QVVVSASDD675 pKa = 3.47GTNSVAQALSITIANVNDD693 pKa = 3.65NAPVITSNGGGGTASLGIAEE713 pKa = 5.14NGTAVTIVTATDD725 pKa = 3.39ADD727 pKa = 4.23GTVPTYY733 pKa = 10.87AIAGGADD740 pKa = 3.06AALFTIDD747 pKa = 4.22PATGALSFLSAPDD760 pKa = 3.82FEE762 pKa = 6.09HH763 pKa = 7.51PLDD766 pKa = 4.46ADD768 pKa = 3.65ANGIYY773 pKa = 9.89QVTVRR778 pKa = 11.84ATDD781 pKa = 3.44GLNVVDD787 pKa = 4.41QLLSVSVTDD796 pKa = 3.47VNEE799 pKa = 3.91VGRR802 pKa = 11.84TITGTSGNNTISPTTTVVGYY822 pKa = 7.47QTTALGDD829 pKa = 3.7TIYY832 pKa = 11.41ALAGNDD838 pKa = 4.05IIDD841 pKa = 4.14GGAGADD847 pKa = 3.9YY848 pKa = 10.48MDD850 pKa = 4.95GGAGNDD856 pKa = 3.35TFYY859 pKa = 11.74VDD861 pKa = 3.46TFSDD865 pKa = 3.66DD866 pKa = 3.9GYY868 pKa = 11.22AGNDD872 pKa = 3.32DD873 pKa = 4.49RR874 pKa = 11.84VIEE877 pKa = 4.03LAGGGTDD884 pKa = 3.68LVYY887 pKa = 11.06ASVSYY892 pKa = 10.8RR893 pKa = 11.84LADD896 pKa = 3.39QVEE899 pKa = 4.18NLTLTGTAALNGYY912 pKa = 10.21GNDD915 pKa = 3.54LANIITGNDD924 pKa = 2.96AANRR928 pKa = 11.84LEE930 pKa = 4.55GGLGADD936 pKa = 3.68TLYY939 pKa = 11.55GMGGNDD945 pKa = 3.7TLDD948 pKa = 3.8GGDD951 pKa = 3.91GNDD954 pKa = 3.41RR955 pKa = 11.84LYY957 pKa = 11.36GGDD960 pKa = 3.73GTDD963 pKa = 3.38TLIGGTGDD971 pKa = 3.49DD972 pKa = 4.27TLDD975 pKa = 3.78GGTGADD981 pKa = 3.6TMTGGDD987 pKa = 4.22GNDD990 pKa = 3.1SYY992 pKa = 12.01YY993 pKa = 11.05VDD995 pKa = 4.2DD996 pKa = 5.85AGDD999 pKa = 3.63QVVEE1003 pKa = 4.18LANGGTTDD1011 pKa = 3.99QIFSAITYY1019 pKa = 7.53SLPAEE1024 pKa = 4.3VEE1026 pKa = 4.14KK1027 pKa = 10.44LTLTGTDD1034 pKa = 3.89AIDD1037 pKa = 3.79GRR1039 pKa = 11.84GNEE1042 pKa = 4.47LNNTITGNGANNGLWGGAGNDD1063 pKa = 3.89TLSGNAGDD1071 pKa = 4.98DD1072 pKa = 3.5RR1073 pKa = 11.84LFGEE1077 pKa = 5.45DD1078 pKa = 5.3GLDD1081 pKa = 3.53TLDD1084 pKa = 4.18GGVGNDD1090 pKa = 3.94RR1091 pKa = 11.84LDD1093 pKa = 3.88GGAGADD1099 pKa = 3.68TLLGKK1104 pKa = 10.31AGNDD1108 pKa = 3.61TLIGGAGKK1116 pKa = 8.54DD1117 pKa = 3.47TLTGGTEE1124 pKa = 3.76ADD1126 pKa = 3.14TFVFNFGDD1134 pKa = 3.64TTLNSASYY1142 pKa = 11.18DD1143 pKa = 3.65RR1144 pKa = 11.84ITDD1147 pKa = 4.24FKK1149 pKa = 11.21ASEE1152 pKa = 4.3GDD1154 pKa = 4.06VIDD1157 pKa = 5.0LDD1159 pKa = 4.22VVNGSLAPADD1169 pKa = 4.04FTATTIATNNFADD1182 pKa = 4.09ALTAAKK1188 pKa = 8.54LTQSAGHH1195 pKa = 5.64ITFVAGATDD1204 pKa = 4.41GWLFWDD1210 pKa = 4.38GNGDD1214 pKa = 3.83GVFDD1218 pKa = 4.41QSVLLSGANTQSAIDD1233 pKa = 3.94PLHH1236 pKa = 6.27IVV1238 pKa = 3.39
Molecular weight: 124.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5E7YES7|A0A5E7YES7_9SPHN 30S ribosomal protein S4 OS=Sphingomonas sp. EC-HK361 OX=2038397 GN=rpsD PE=3 SV=1
MM1 pKa = 7.59ARR3 pKa = 11.84TSLPYY8 pKa = 10.37RR9 pKa = 11.84ITAQALQSRR18 pKa = 11.84ARR20 pKa = 11.84APRR23 pKa = 11.84AARR26 pKa = 11.84SISAARR32 pKa = 11.84VVFAPRR38 pKa = 11.84LAVGRR43 pKa = 11.84PGSS46 pKa = 3.44
MM1 pKa = 7.59ARR3 pKa = 11.84TSLPYY8 pKa = 10.37RR9 pKa = 11.84ITAQALQSRR18 pKa = 11.84ARR20 pKa = 11.84APRR23 pKa = 11.84AARR26 pKa = 11.84SISAARR32 pKa = 11.84VVFAPRR38 pKa = 11.84LAVGRR43 pKa = 11.84PGSS46 pKa = 3.44
Molecular weight: 4.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1014211 |
20 |
1842 |
311.5 |
33.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.165 ± 0.082 | 0.75 ± 0.015 |
6.172 ± 0.031 | 4.909 ± 0.04 |
3.462 ± 0.028 | 8.975 ± 0.041 |
1.946 ± 0.026 | 4.977 ± 0.025 |
2.872 ± 0.037 | 9.581 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.356 ± 0.021 | 2.375 ± 0.032 |
5.395 ± 0.033 | 2.951 ± 0.028 |
7.604 ± 0.049 | 4.964 ± 0.036 |
5.609 ± 0.038 | 7.348 ± 0.033 |
1.407 ± 0.019 | 2.183 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
