
Methanobacterium lacus (strain AL-21)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanomada group; Methanobacteria; Methanobacteriales; Methanobacteriaceae; Methanobacterium
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
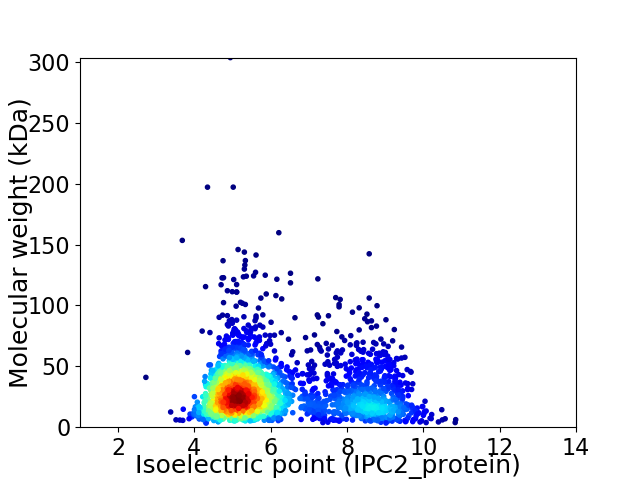
Virtual 2D-PAGE plot for 2493 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0T7P3|F0T7P3_METLA Uncharacterized protein OS=Methanobacterium lacus (strain AL-21) OX=877455 GN=Metbo_1371 PE=4 SV=1
MM1 pKa = 8.26VIMAFMFFGILSTSSAANMTGTWSGSNGVYY31 pKa = 10.56NSTAGPVKK39 pKa = 10.74VNFTEE44 pKa = 4.08KK45 pKa = 10.58HH46 pKa = 5.22NGAATVTATPTGTTNTNTAFWSNASAAGHH75 pKa = 6.11SSLEE79 pKa = 4.28VTLSWNTLTKK89 pKa = 10.73ANNGTITFNFNRR101 pKa = 11.84PVNNPILYY109 pKa = 9.43IDD111 pKa = 4.85RR112 pKa = 11.84IGGNLQNGGTITSSSALLTLLNSGMYY138 pKa = 8.25LTKK141 pKa = 10.84LSGPTSFEE149 pKa = 4.24VTSSTIQRR157 pKa = 11.84TPGVTISSTSTEE169 pKa = 3.67ASMDD173 pKa = 4.0PLGGTAAGTVQIMGTNITSVTFSWWGVGADD203 pKa = 3.64SQADD207 pKa = 3.5GLEE210 pKa = 4.17FLWEE214 pKa = 3.95LEE216 pKa = 4.12APVDD220 pKa = 4.23LSISKK225 pKa = 10.19SDD227 pKa = 3.59SPDD230 pKa = 3.24PVVAGKK236 pKa = 9.98NLTYY240 pKa = 10.49TITVHH245 pKa = 6.35NNGPSSIISSDD256 pKa = 3.33VFTVIDD262 pKa = 3.95NLPAGFTATSYY273 pKa = 10.37TPSEE277 pKa = 4.03GSYY280 pKa = 11.18NPVTGAWTGVTLANGEE296 pKa = 4.47DD297 pKa = 3.52VTLTIIGIVNSTKK310 pKa = 10.43TGTLTNTASVQVPSGITDD328 pKa = 3.81PNTNNNQAPPVITTVDD344 pKa = 3.68GLPTANNDD352 pKa = 3.38SKK354 pKa = 8.73TTPEE358 pKa = 4.09DD359 pKa = 3.75TPISGNLPASDD370 pKa = 4.0PDD372 pKa = 4.0GPVTVVNFVINGTTYY387 pKa = 11.26APGTVNIPGVGKK399 pKa = 10.27IVINSDD405 pKa = 2.54GSYY408 pKa = 10.94LFTPASNYY416 pKa = 9.78NGPVPTITYY425 pKa = 7.28TASDD429 pKa = 3.65NTGNTATGNLNINVTPVNDD448 pKa = 3.86APVGTGDD455 pKa = 4.61SKK457 pKa = 8.75TTPEE461 pKa = 3.97DD462 pKa = 3.41TQATGKK468 pKa = 8.41VTGTDD473 pKa = 3.0VDD475 pKa = 4.0GDD477 pKa = 3.99TLTFAKK483 pKa = 9.77DD484 pKa = 3.27TDD486 pKa = 4.24PSHH489 pKa = 5.82GTVVVNADD497 pKa = 3.01GTYY500 pKa = 10.23TYY502 pKa = 10.12MPNANYY508 pKa = 10.24NGPDD512 pKa = 3.33SFTVIVSDD520 pKa = 4.0GKK522 pKa = 10.77GGSDD526 pKa = 2.49IVTVNINVTPVNDD539 pKa = 3.73APIGNGDD546 pKa = 3.84SKK548 pKa = 9.92TIPEE552 pKa = 4.21DD553 pKa = 3.53TQATGKK559 pKa = 8.41VTGTDD564 pKa = 2.89VDD566 pKa = 4.35GDD568 pKa = 3.43ILTYY572 pKa = 10.02TKK574 pKa = 10.89NSNPTHH580 pKa = 5.37GTVVVNADD588 pKa = 3.01GTYY591 pKa = 10.23TYY593 pKa = 10.12MPNANYY599 pKa = 10.24NGPDD603 pKa = 3.33SFTVIVSDD611 pKa = 4.43GNGGTDD617 pKa = 2.95TVTVTINVTPVNDD630 pKa = 3.81APIGTGDD637 pKa = 3.51SKK639 pKa = 11.42AIPEE643 pKa = 4.13DD644 pKa = 3.76TQATGKK650 pKa = 8.41VTGTDD655 pKa = 3.0VDD657 pKa = 4.43GDD659 pKa = 3.9TLTYY663 pKa = 9.7TKK665 pKa = 10.8NSNPTHH671 pKa = 5.35GTAVVNADD679 pKa = 2.95GTYY682 pKa = 10.08TYY684 pKa = 10.37TPNANYY690 pKa = 10.2NGPDD694 pKa = 3.12SFTIIVSDD702 pKa = 4.02GNGGSDD708 pKa = 3.45VVTVTIDD715 pKa = 3.28VTPVNDD721 pKa = 3.79APVGTGDD728 pKa = 4.02SKK730 pKa = 10.0TIPEE734 pKa = 4.21DD735 pKa = 3.53TQATGKK741 pKa = 8.41VTGTDD746 pKa = 3.0VDD748 pKa = 4.43GDD750 pKa = 3.9TLTYY754 pKa = 9.88TKK756 pKa = 9.54NTNPSHH762 pKa = 5.38GTAVVNADD770 pKa = 2.95GTYY773 pKa = 10.08TYY775 pKa = 10.37TPNANYY781 pKa = 10.2NGPDD785 pKa = 3.35SFTVTVSDD793 pKa = 4.17GNGGSDD799 pKa = 3.45VVTVTIDD806 pKa = 3.28VTPVNDD812 pKa = 3.79APVGTGDD819 pKa = 4.02SKK821 pKa = 10.0TIPEE825 pKa = 4.21DD826 pKa = 3.53TQATGKK832 pKa = 8.41VTGTDD837 pKa = 3.0VDD839 pKa = 4.43GDD841 pKa = 3.9TLTYY845 pKa = 9.92TKK847 pKa = 9.83NTDD850 pKa = 3.54PSHH853 pKa = 5.44GTAVVNADD861 pKa = 2.95GTYY864 pKa = 10.08TYY866 pKa = 10.37TPNANYY872 pKa = 10.2NGPDD876 pKa = 3.35SFTVTVSDD884 pKa = 4.39GNGGTDD890 pKa = 2.95TVTVTINVTPVNDD903 pKa = 3.76APIGNDD909 pKa = 3.26DD910 pKa = 4.22SKK912 pKa = 10.14TIPEE916 pKa = 4.17DD917 pKa = 3.53TQATGKK923 pKa = 8.41VTGTDD928 pKa = 3.0VDD930 pKa = 4.43GDD932 pKa = 3.9TLTYY936 pKa = 9.63TKK938 pKa = 10.78NSNPSHH944 pKa = 5.45GTAVVNADD952 pKa = 2.95GTYY955 pKa = 10.08TYY957 pKa = 10.37TPNANYY963 pKa = 10.2NGPDD967 pKa = 3.12SFTIIVSDD975 pKa = 3.79GKK977 pKa = 11.02GGSDD981 pKa = 3.18VVTVNINVTPVNDD994 pKa = 3.76APIGNDD1000 pKa = 3.26DD1001 pKa = 4.22SKK1003 pKa = 10.14TIPEE1007 pKa = 4.19DD1008 pKa = 3.55TQATGNVTGTDD1019 pKa = 3.17VDD1021 pKa = 4.38GDD1023 pKa = 3.89TLTYY1027 pKa = 9.93TKK1029 pKa = 9.99STDD1032 pKa = 3.57PSHH1035 pKa = 5.75GTAVVNADD1043 pKa = 2.95GTYY1046 pKa = 10.08TYY1048 pKa = 10.37TPNANYY1054 pKa = 10.2NGPDD1058 pKa = 3.35SFTVTVSDD1066 pKa = 4.17GNGGSDD1072 pKa = 3.26VVTVNINVTPVNDD1085 pKa = 3.76APIGNDD1091 pKa = 3.26DD1092 pKa = 4.22SKK1094 pKa = 10.14TIPEE1098 pKa = 4.17DD1099 pKa = 3.53TQATGKK1105 pKa = 8.4VTGTDD1110 pKa = 2.83IDD1112 pKa = 4.74GDD1114 pKa = 3.93TLTYY1118 pKa = 9.92TKK1120 pKa = 9.54NTNPSHH1126 pKa = 5.38GTAVVNADD1134 pKa = 2.95GTYY1137 pKa = 10.08TYY1139 pKa = 10.37TPNANYY1145 pKa = 10.2NGPDD1149 pKa = 3.35SFTVTVSDD1157 pKa = 4.17GNGGSDD1163 pKa = 3.45VVTVTIDD1170 pKa = 3.19VMPVNDD1176 pKa = 4.41APVGTGDD1183 pKa = 3.61SQTIPEE1189 pKa = 4.38DD1190 pKa = 3.58TQATGKK1196 pKa = 8.41VTGTDD1201 pKa = 3.0VDD1203 pKa = 4.43GDD1205 pKa = 3.9TLTYY1209 pKa = 9.92TKK1211 pKa = 9.83NTDD1214 pKa = 3.54PSHH1217 pKa = 5.44GTAVVNADD1225 pKa = 2.95GTYY1228 pKa = 10.08TYY1230 pKa = 10.37TPNANYY1236 pKa = 10.2NGPDD1240 pKa = 3.35SFTVTVSDD1248 pKa = 4.17GNGGSDD1254 pKa = 3.45VVTVTIDD1261 pKa = 3.28VTPVNDD1267 pKa = 3.79APVGTGDD1274 pKa = 4.02SKK1276 pKa = 10.0TIPEE1280 pKa = 4.21DD1281 pKa = 3.53TQATGKK1287 pKa = 10.92VIGTDD1292 pKa = 3.09VDD1294 pKa = 4.37GDD1296 pKa = 3.89TLTYY1300 pKa = 9.63TKK1302 pKa = 10.78NSNPSHH1308 pKa = 5.45GTAVVNADD1316 pKa = 2.95GTYY1319 pKa = 10.08TYY1321 pKa = 10.37TPNANYY1327 pKa = 10.2NGPDD1331 pKa = 3.12SFTIIVSDD1339 pKa = 3.79GKK1341 pKa = 11.02GGSDD1345 pKa = 3.41VVTVTIDD1352 pKa = 3.28VTPVNDD1358 pKa = 3.79APVGTGDD1365 pKa = 3.61SQTIPEE1371 pKa = 4.38DD1372 pKa = 3.58TQATGKK1378 pKa = 8.41VTGTDD1383 pKa = 3.0VDD1385 pKa = 4.43GDD1387 pKa = 3.9TLTYY1391 pKa = 9.91TKK1393 pKa = 9.55NTNPTHH1399 pKa = 5.27GTVVVNADD1407 pKa = 3.0GTYY1410 pKa = 10.08TYY1412 pKa = 10.37TPNANYY1418 pKa = 10.2NGPDD1422 pKa = 3.12SFTIIVSDD1430 pKa = 3.79GKK1432 pKa = 11.04GGSDD1436 pKa = 3.1VVLVNITMTPVDD1448 pKa = 4.44DD1449 pKa = 3.96PVKK1452 pKa = 10.68NVTPTKK1458 pKa = 10.55VKK1460 pKa = 10.36NQTEE1464 pKa = 4.33VGTIPMQRR1472 pKa = 11.84TGVPLPMLFMALVMIFGAFVSNKK1495 pKa = 9.38RR1496 pKa = 11.84KK1497 pKa = 9.99
MM1 pKa = 8.26VIMAFMFFGILSTSSAANMTGTWSGSNGVYY31 pKa = 10.56NSTAGPVKK39 pKa = 10.74VNFTEE44 pKa = 4.08KK45 pKa = 10.58HH46 pKa = 5.22NGAATVTATPTGTTNTNTAFWSNASAAGHH75 pKa = 6.11SSLEE79 pKa = 4.28VTLSWNTLTKK89 pKa = 10.73ANNGTITFNFNRR101 pKa = 11.84PVNNPILYY109 pKa = 9.43IDD111 pKa = 4.85RR112 pKa = 11.84IGGNLQNGGTITSSSALLTLLNSGMYY138 pKa = 8.25LTKK141 pKa = 10.84LSGPTSFEE149 pKa = 4.24VTSSTIQRR157 pKa = 11.84TPGVTISSTSTEE169 pKa = 3.67ASMDD173 pKa = 4.0PLGGTAAGTVQIMGTNITSVTFSWWGVGADD203 pKa = 3.64SQADD207 pKa = 3.5GLEE210 pKa = 4.17FLWEE214 pKa = 3.95LEE216 pKa = 4.12APVDD220 pKa = 4.23LSISKK225 pKa = 10.19SDD227 pKa = 3.59SPDD230 pKa = 3.24PVVAGKK236 pKa = 9.98NLTYY240 pKa = 10.49TITVHH245 pKa = 6.35NNGPSSIISSDD256 pKa = 3.33VFTVIDD262 pKa = 3.95NLPAGFTATSYY273 pKa = 10.37TPSEE277 pKa = 4.03GSYY280 pKa = 11.18NPVTGAWTGVTLANGEE296 pKa = 4.47DD297 pKa = 3.52VTLTIIGIVNSTKK310 pKa = 10.43TGTLTNTASVQVPSGITDD328 pKa = 3.81PNTNNNQAPPVITTVDD344 pKa = 3.68GLPTANNDD352 pKa = 3.38SKK354 pKa = 8.73TTPEE358 pKa = 4.09DD359 pKa = 3.75TPISGNLPASDD370 pKa = 4.0PDD372 pKa = 4.0GPVTVVNFVINGTTYY387 pKa = 11.26APGTVNIPGVGKK399 pKa = 10.27IVINSDD405 pKa = 2.54GSYY408 pKa = 10.94LFTPASNYY416 pKa = 9.78NGPVPTITYY425 pKa = 7.28TASDD429 pKa = 3.65NTGNTATGNLNINVTPVNDD448 pKa = 3.86APVGTGDD455 pKa = 4.61SKK457 pKa = 8.75TTPEE461 pKa = 3.97DD462 pKa = 3.41TQATGKK468 pKa = 8.41VTGTDD473 pKa = 3.0VDD475 pKa = 4.0GDD477 pKa = 3.99TLTFAKK483 pKa = 9.77DD484 pKa = 3.27TDD486 pKa = 4.24PSHH489 pKa = 5.82GTVVVNADD497 pKa = 3.01GTYY500 pKa = 10.23TYY502 pKa = 10.12MPNANYY508 pKa = 10.24NGPDD512 pKa = 3.33SFTVIVSDD520 pKa = 4.0GKK522 pKa = 10.77GGSDD526 pKa = 2.49IVTVNINVTPVNDD539 pKa = 3.73APIGNGDD546 pKa = 3.84SKK548 pKa = 9.92TIPEE552 pKa = 4.21DD553 pKa = 3.53TQATGKK559 pKa = 8.41VTGTDD564 pKa = 2.89VDD566 pKa = 4.35GDD568 pKa = 3.43ILTYY572 pKa = 10.02TKK574 pKa = 10.89NSNPTHH580 pKa = 5.37GTVVVNADD588 pKa = 3.01GTYY591 pKa = 10.23TYY593 pKa = 10.12MPNANYY599 pKa = 10.24NGPDD603 pKa = 3.33SFTVIVSDD611 pKa = 4.43GNGGTDD617 pKa = 2.95TVTVTINVTPVNDD630 pKa = 3.81APIGTGDD637 pKa = 3.51SKK639 pKa = 11.42AIPEE643 pKa = 4.13DD644 pKa = 3.76TQATGKK650 pKa = 8.41VTGTDD655 pKa = 3.0VDD657 pKa = 4.43GDD659 pKa = 3.9TLTYY663 pKa = 9.7TKK665 pKa = 10.8NSNPTHH671 pKa = 5.35GTAVVNADD679 pKa = 2.95GTYY682 pKa = 10.08TYY684 pKa = 10.37TPNANYY690 pKa = 10.2NGPDD694 pKa = 3.12SFTIIVSDD702 pKa = 4.02GNGGSDD708 pKa = 3.45VVTVTIDD715 pKa = 3.28VTPVNDD721 pKa = 3.79APVGTGDD728 pKa = 4.02SKK730 pKa = 10.0TIPEE734 pKa = 4.21DD735 pKa = 3.53TQATGKK741 pKa = 8.41VTGTDD746 pKa = 3.0VDD748 pKa = 4.43GDD750 pKa = 3.9TLTYY754 pKa = 9.88TKK756 pKa = 9.54NTNPSHH762 pKa = 5.38GTAVVNADD770 pKa = 2.95GTYY773 pKa = 10.08TYY775 pKa = 10.37TPNANYY781 pKa = 10.2NGPDD785 pKa = 3.35SFTVTVSDD793 pKa = 4.17GNGGSDD799 pKa = 3.45VVTVTIDD806 pKa = 3.28VTPVNDD812 pKa = 3.79APVGTGDD819 pKa = 4.02SKK821 pKa = 10.0TIPEE825 pKa = 4.21DD826 pKa = 3.53TQATGKK832 pKa = 8.41VTGTDD837 pKa = 3.0VDD839 pKa = 4.43GDD841 pKa = 3.9TLTYY845 pKa = 9.92TKK847 pKa = 9.83NTDD850 pKa = 3.54PSHH853 pKa = 5.44GTAVVNADD861 pKa = 2.95GTYY864 pKa = 10.08TYY866 pKa = 10.37TPNANYY872 pKa = 10.2NGPDD876 pKa = 3.35SFTVTVSDD884 pKa = 4.39GNGGTDD890 pKa = 2.95TVTVTINVTPVNDD903 pKa = 3.76APIGNDD909 pKa = 3.26DD910 pKa = 4.22SKK912 pKa = 10.14TIPEE916 pKa = 4.17DD917 pKa = 3.53TQATGKK923 pKa = 8.41VTGTDD928 pKa = 3.0VDD930 pKa = 4.43GDD932 pKa = 3.9TLTYY936 pKa = 9.63TKK938 pKa = 10.78NSNPSHH944 pKa = 5.45GTAVVNADD952 pKa = 2.95GTYY955 pKa = 10.08TYY957 pKa = 10.37TPNANYY963 pKa = 10.2NGPDD967 pKa = 3.12SFTIIVSDD975 pKa = 3.79GKK977 pKa = 11.02GGSDD981 pKa = 3.18VVTVNINVTPVNDD994 pKa = 3.76APIGNDD1000 pKa = 3.26DD1001 pKa = 4.22SKK1003 pKa = 10.14TIPEE1007 pKa = 4.19DD1008 pKa = 3.55TQATGNVTGTDD1019 pKa = 3.17VDD1021 pKa = 4.38GDD1023 pKa = 3.89TLTYY1027 pKa = 9.93TKK1029 pKa = 9.99STDD1032 pKa = 3.57PSHH1035 pKa = 5.75GTAVVNADD1043 pKa = 2.95GTYY1046 pKa = 10.08TYY1048 pKa = 10.37TPNANYY1054 pKa = 10.2NGPDD1058 pKa = 3.35SFTVTVSDD1066 pKa = 4.17GNGGSDD1072 pKa = 3.26VVTVNINVTPVNDD1085 pKa = 3.76APIGNDD1091 pKa = 3.26DD1092 pKa = 4.22SKK1094 pKa = 10.14TIPEE1098 pKa = 4.17DD1099 pKa = 3.53TQATGKK1105 pKa = 8.4VTGTDD1110 pKa = 2.83IDD1112 pKa = 4.74GDD1114 pKa = 3.93TLTYY1118 pKa = 9.92TKK1120 pKa = 9.54NTNPSHH1126 pKa = 5.38GTAVVNADD1134 pKa = 2.95GTYY1137 pKa = 10.08TYY1139 pKa = 10.37TPNANYY1145 pKa = 10.2NGPDD1149 pKa = 3.35SFTVTVSDD1157 pKa = 4.17GNGGSDD1163 pKa = 3.45VVTVTIDD1170 pKa = 3.19VMPVNDD1176 pKa = 4.41APVGTGDD1183 pKa = 3.61SQTIPEE1189 pKa = 4.38DD1190 pKa = 3.58TQATGKK1196 pKa = 8.41VTGTDD1201 pKa = 3.0VDD1203 pKa = 4.43GDD1205 pKa = 3.9TLTYY1209 pKa = 9.92TKK1211 pKa = 9.83NTDD1214 pKa = 3.54PSHH1217 pKa = 5.44GTAVVNADD1225 pKa = 2.95GTYY1228 pKa = 10.08TYY1230 pKa = 10.37TPNANYY1236 pKa = 10.2NGPDD1240 pKa = 3.35SFTVTVSDD1248 pKa = 4.17GNGGSDD1254 pKa = 3.45VVTVTIDD1261 pKa = 3.28VTPVNDD1267 pKa = 3.79APVGTGDD1274 pKa = 4.02SKK1276 pKa = 10.0TIPEE1280 pKa = 4.21DD1281 pKa = 3.53TQATGKK1287 pKa = 10.92VIGTDD1292 pKa = 3.09VDD1294 pKa = 4.37GDD1296 pKa = 3.89TLTYY1300 pKa = 9.63TKK1302 pKa = 10.78NSNPSHH1308 pKa = 5.45GTAVVNADD1316 pKa = 2.95GTYY1319 pKa = 10.08TYY1321 pKa = 10.37TPNANYY1327 pKa = 10.2NGPDD1331 pKa = 3.12SFTIIVSDD1339 pKa = 3.79GKK1341 pKa = 11.02GGSDD1345 pKa = 3.41VVTVTIDD1352 pKa = 3.28VTPVNDD1358 pKa = 3.79APVGTGDD1365 pKa = 3.61SQTIPEE1371 pKa = 4.38DD1372 pKa = 3.58TQATGKK1378 pKa = 8.41VTGTDD1383 pKa = 3.0VDD1385 pKa = 4.43GDD1387 pKa = 3.9TLTYY1391 pKa = 9.91TKK1393 pKa = 9.55NTNPTHH1399 pKa = 5.27GTVVVNADD1407 pKa = 3.0GTYY1410 pKa = 10.08TYY1412 pKa = 10.37TPNANYY1418 pKa = 10.2NGPDD1422 pKa = 3.12SFTIIVSDD1430 pKa = 3.79GKK1432 pKa = 11.04GGSDD1436 pKa = 3.1VVLVNITMTPVDD1448 pKa = 4.44DD1449 pKa = 3.96PVKK1452 pKa = 10.68NVTPTKK1458 pKa = 10.55VKK1460 pKa = 10.36NQTEE1464 pKa = 4.33VGTIPMQRR1472 pKa = 11.84TGVPLPMLFMALVMIFGAFVSNKK1495 pKa = 9.38RR1496 pKa = 11.84KK1497 pKa = 9.99
Molecular weight: 153.38 kDa
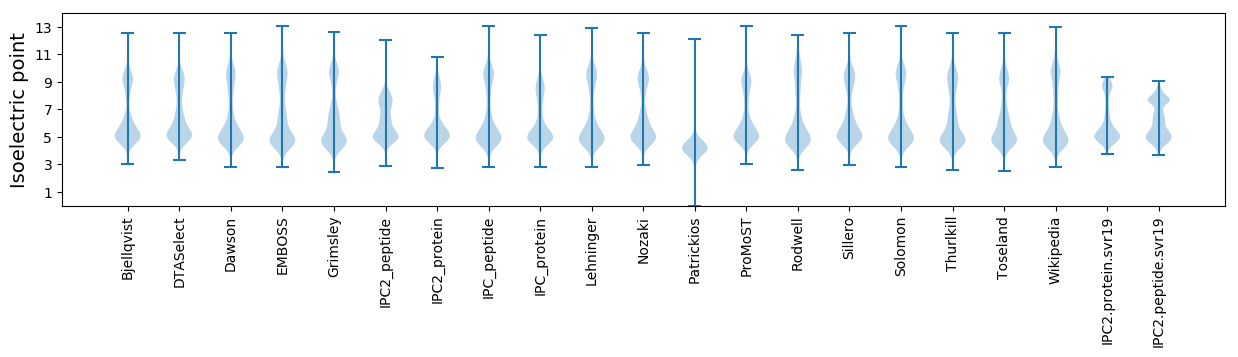
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0TA09|F0TA09_METLA Isopropylmalate/isohomocitrate dehydrogenase OS=Methanobacterium lacus (strain AL-21) OX=877455 GN=Metbo_0581 PE=3 SV=1
MM1 pKa = 7.46SRR3 pKa = 11.84NKK5 pKa = 9.93PCAKK9 pKa = 10.18KK10 pKa = 10.51LRR12 pKa = 11.84LSKK15 pKa = 9.8ATKK18 pKa = 8.18QNRR21 pKa = 11.84RR22 pKa = 11.84VPLWVMLKK30 pKa = 9.12TSRR33 pKa = 11.84KK34 pKa = 9.07VRR36 pKa = 11.84THH38 pKa = 5.73PKK40 pKa = 7.11MRR42 pKa = 11.84QWRR45 pKa = 11.84RR46 pKa = 11.84SKK48 pKa = 11.06VKK50 pKa = 10.52AA51 pKa = 3.54
MM1 pKa = 7.46SRR3 pKa = 11.84NKK5 pKa = 9.93PCAKK9 pKa = 10.18KK10 pKa = 10.51LRR12 pKa = 11.84LSKK15 pKa = 9.8ATKK18 pKa = 8.18QNRR21 pKa = 11.84RR22 pKa = 11.84VPLWVMLKK30 pKa = 9.12TSRR33 pKa = 11.84KK34 pKa = 9.07VRR36 pKa = 11.84THH38 pKa = 5.73PKK40 pKa = 7.11MRR42 pKa = 11.84QWRR45 pKa = 11.84RR46 pKa = 11.84SKK48 pKa = 11.06VKK50 pKa = 10.52AA51 pKa = 3.54
Molecular weight: 6.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
715694 |
30 |
2864 |
287.1 |
32.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.074 ± 0.06 | 1.125 ± 0.028 |
5.601 ± 0.045 | 6.956 ± 0.079 |
4.145 ± 0.042 | 6.996 ± 0.058 |
1.691 ± 0.02 | 8.915 ± 0.054 |
7.483 ± 0.057 | 9.097 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.769 ± 0.032 | 5.752 ± 0.093 |
3.631 ± 0.03 | 2.487 ± 0.023 |
3.383 ± 0.044 | 6.547 ± 0.046 |
5.776 ± 0.085 | 7.175 ± 0.051 |
0.794 ± 0.018 | 3.602 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
