
Alkalibaculum bacchi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Alkalibaculum
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
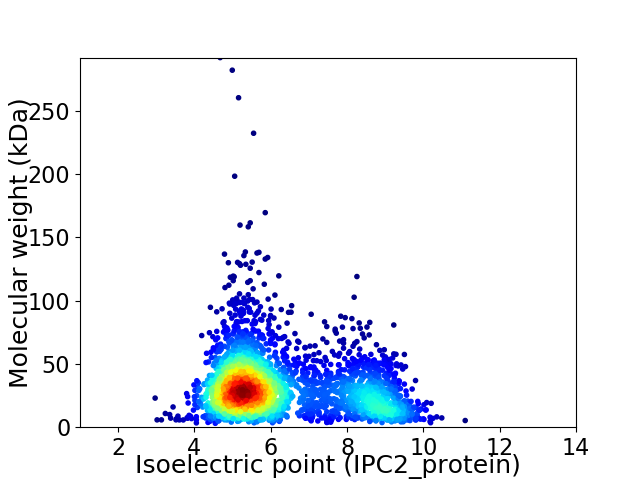
Virtual 2D-PAGE plot for 2857 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
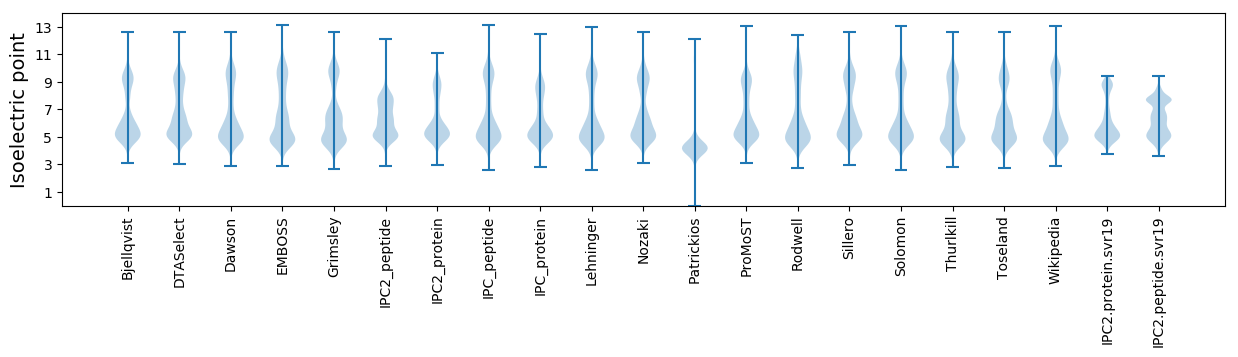
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A366IC42|A0A366IC42_9FIRM Putative alkaline shock family protein YloU OS=Alkalibaculum bacchi OX=645887 GN=DES36_10351 PE=3 SV=1
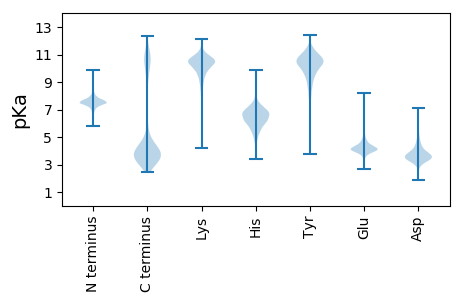
MM1 pKa = 7.4KK2 pKa = 10.26KK3 pKa = 10.42VIAFCMVVFMVFTLCACKK21 pKa = 10.15PKK23 pKa = 10.77GDD25 pKa = 3.89EE26 pKa = 4.39QKK28 pKa = 11.11DD29 pKa = 3.53KK30 pKa = 11.48QDD32 pKa = 3.46INEE35 pKa = 4.24SSDD38 pKa = 3.06GRR40 pKa = 11.84GDD42 pKa = 3.45VFDD45 pKa = 5.39DD46 pKa = 3.66YY47 pKa = 11.17KK48 pKa = 10.9IPEE51 pKa = 4.48GADD54 pKa = 3.61EE55 pKa = 3.9IRR57 pKa = 11.84QGVFYY62 pKa = 11.26YY63 pKa = 10.43EE64 pKa = 4.52LYY66 pKa = 10.37EE67 pKa = 4.34LSGDD71 pKa = 3.99PGDD74 pKa = 3.94GLTPRR79 pKa = 11.84EE80 pKa = 4.1GAGLVDD86 pKa = 4.61EE87 pKa = 5.38LLASTGLYY95 pKa = 10.27SGDD98 pKa = 3.71RR99 pKa = 11.84ISADD103 pKa = 2.98EE104 pKa = 4.14CLYY107 pKa = 10.66ISFDD111 pKa = 3.91DD112 pKa = 5.44LMIIDD117 pKa = 3.84SAMGRR122 pKa = 11.84EE123 pKa = 4.21CYY125 pKa = 9.72IYY127 pKa = 10.85SVALGTAEE135 pKa = 4.36GGLMGDD141 pKa = 4.86GYY143 pKa = 10.31QVIYY147 pKa = 10.11RR148 pKa = 11.84FSVDD152 pKa = 3.13YY153 pKa = 11.0SGNKK157 pKa = 7.31TASIYY162 pKa = 10.91EE163 pKa = 4.34DD164 pKa = 4.03FSDD167 pKa = 4.9TILDD171 pKa = 4.29DD172 pKa = 3.99GLGDD176 pKa = 4.84VISKK180 pKa = 11.05DD181 pKa = 3.34NTHH184 pKa = 6.81TDD186 pKa = 3.17NTSKK190 pKa = 11.15DD191 pKa = 3.56DD192 pKa = 3.54GRR194 pKa = 11.84GDD196 pKa = 4.1VLSQDD201 pKa = 4.08DD202 pKa = 4.54GKK204 pKa = 11.72GDD206 pKa = 5.23LIPYY210 pKa = 8.03GPNWRR215 pKa = 11.84GMYY218 pKa = 10.34VGDD221 pKa = 4.07EE222 pKa = 4.07FSIEE226 pKa = 3.61ITKK229 pKa = 10.48FNGSSFWFDD238 pKa = 2.46IYY240 pKa = 11.14LLRR243 pKa = 11.84NGSTVMSGTATLYY256 pKa = 10.49PDD258 pKa = 4.16DD259 pKa = 5.63KK260 pKa = 11.86YY261 pKa = 10.09MAEE264 pKa = 3.95YY265 pKa = 11.16GEE267 pKa = 4.77ISFSLYY273 pKa = 10.46KK274 pKa = 10.43DD275 pKa = 3.27YY276 pKa = 11.56SAIDD280 pKa = 3.51FFAPEE285 pKa = 4.08SSDD288 pKa = 3.12WAHH291 pKa = 6.28MRR293 pKa = 11.84GQYY296 pKa = 10.85KK297 pKa = 10.32FIEE300 pKa = 4.32
MM1 pKa = 7.4KK2 pKa = 10.26KK3 pKa = 10.42VIAFCMVVFMVFTLCACKK21 pKa = 10.15PKK23 pKa = 10.77GDD25 pKa = 3.89EE26 pKa = 4.39QKK28 pKa = 11.11DD29 pKa = 3.53KK30 pKa = 11.48QDD32 pKa = 3.46INEE35 pKa = 4.24SSDD38 pKa = 3.06GRR40 pKa = 11.84GDD42 pKa = 3.45VFDD45 pKa = 5.39DD46 pKa = 3.66YY47 pKa = 11.17KK48 pKa = 10.9IPEE51 pKa = 4.48GADD54 pKa = 3.61EE55 pKa = 3.9IRR57 pKa = 11.84QGVFYY62 pKa = 11.26YY63 pKa = 10.43EE64 pKa = 4.52LYY66 pKa = 10.37EE67 pKa = 4.34LSGDD71 pKa = 3.99PGDD74 pKa = 3.94GLTPRR79 pKa = 11.84EE80 pKa = 4.1GAGLVDD86 pKa = 4.61EE87 pKa = 5.38LLASTGLYY95 pKa = 10.27SGDD98 pKa = 3.71RR99 pKa = 11.84ISADD103 pKa = 2.98EE104 pKa = 4.14CLYY107 pKa = 10.66ISFDD111 pKa = 3.91DD112 pKa = 5.44LMIIDD117 pKa = 3.84SAMGRR122 pKa = 11.84EE123 pKa = 4.21CYY125 pKa = 9.72IYY127 pKa = 10.85SVALGTAEE135 pKa = 4.36GGLMGDD141 pKa = 4.86GYY143 pKa = 10.31QVIYY147 pKa = 10.11RR148 pKa = 11.84FSVDD152 pKa = 3.13YY153 pKa = 11.0SGNKK157 pKa = 7.31TASIYY162 pKa = 10.91EE163 pKa = 4.34DD164 pKa = 4.03FSDD167 pKa = 4.9TILDD171 pKa = 4.29DD172 pKa = 3.99GLGDD176 pKa = 4.84VISKK180 pKa = 11.05DD181 pKa = 3.34NTHH184 pKa = 6.81TDD186 pKa = 3.17NTSKK190 pKa = 11.15DD191 pKa = 3.56DD192 pKa = 3.54GRR194 pKa = 11.84GDD196 pKa = 4.1VLSQDD201 pKa = 4.08DD202 pKa = 4.54GKK204 pKa = 11.72GDD206 pKa = 5.23LIPYY210 pKa = 8.03GPNWRR215 pKa = 11.84GMYY218 pKa = 10.34VGDD221 pKa = 4.07EE222 pKa = 4.07FSIEE226 pKa = 3.61ITKK229 pKa = 10.48FNGSSFWFDD238 pKa = 2.46IYY240 pKa = 11.14LLRR243 pKa = 11.84NGSTVMSGTATLYY256 pKa = 10.49PDD258 pKa = 4.16DD259 pKa = 5.63KK260 pKa = 11.86YY261 pKa = 10.09MAEE264 pKa = 3.95YY265 pKa = 11.16GEE267 pKa = 4.77ISFSLYY273 pKa = 10.46KK274 pKa = 10.43DD275 pKa = 3.27YY276 pKa = 11.56SAIDD280 pKa = 3.51FFAPEE285 pKa = 4.08SSDD288 pKa = 3.12WAHH291 pKa = 6.28MRR293 pKa = 11.84GQYY296 pKa = 10.85KK297 pKa = 10.32FIEE300 pKa = 4.32
Molecular weight: 33.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A366I3J4|A0A366I3J4_9FIRM Ribulose-phosphate 3-epimerase OS=Alkalibaculum bacchi OX=645887 GN=DES36_11420 PE=3 SV=1
MM1 pKa = 7.14KK2 pKa = 9.31QTYY5 pKa = 7.67QPKK8 pKa = 8.45KK9 pKa = 7.59RR10 pKa = 11.84KK11 pKa = 9.02RR12 pKa = 11.84SRR14 pKa = 11.84VHH16 pKa = 6.41GFRR19 pKa = 11.84KK20 pKa = 10.02RR21 pKa = 11.84MASSTGQNVLSNRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.97GRR39 pKa = 11.84KK40 pKa = 8.84RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.14KK2 pKa = 9.31QTYY5 pKa = 7.67QPKK8 pKa = 8.45KK9 pKa = 7.59RR10 pKa = 11.84KK11 pKa = 9.02RR12 pKa = 11.84SRR14 pKa = 11.84VHH16 pKa = 6.41GFRR19 pKa = 11.84KK20 pKa = 10.02RR21 pKa = 11.84MASSTGQNVLSNRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.97GRR39 pKa = 11.84KK40 pKa = 8.84RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
853553 |
29 |
2535 |
298.8 |
33.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.95 ± 0.051 | 1.134 ± 0.02 |
5.576 ± 0.033 | 7.611 ± 0.054 |
4.214 ± 0.039 | 6.592 ± 0.049 |
1.735 ± 0.021 | 9.437 ± 0.051 |
8.009 ± 0.046 | 9.247 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.756 ± 0.023 | 5.288 ± 0.036 |
3.102 ± 0.027 | 3.12 ± 0.024 |
3.675 ± 0.03 | 6.214 ± 0.036 |
5.083 ± 0.03 | 6.522 ± 0.035 |
0.704 ± 0.014 | 4.034 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
