
Gordonia phage CloverMinnie
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Sourvirus; unclassified Sourvirus
Average proteome isoelectric point is 5.6
Get precalculated fractions of proteins
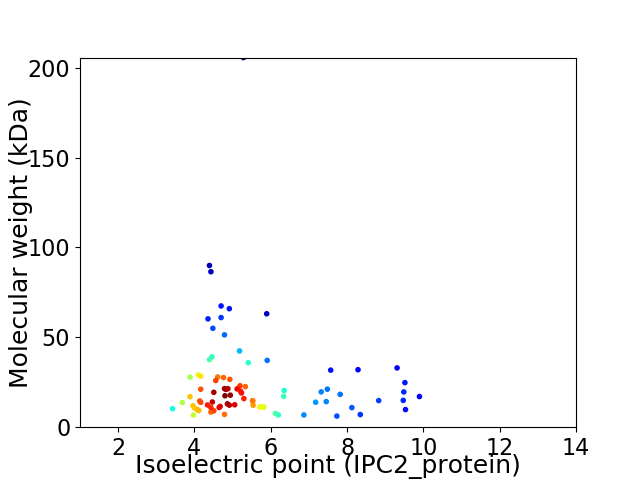
Virtual 2D-PAGE plot for 84 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5J6TKD1|A0A5J6TKD1_9CAUD Uncharacterized protein OS=Gordonia phage CloverMinnie OX=2599842 GN=83 PE=4 SV=1
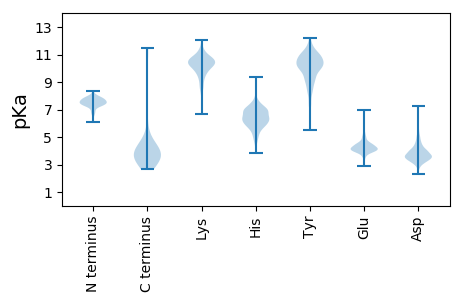
MM1 pKa = 7.38ATPTFEE7 pKa = 4.0PTTLRR12 pKa = 11.84GHH14 pKa = 6.5ALLRR18 pKa = 11.84HH19 pKa = 4.79VLEE22 pKa = 5.39IIEE25 pKa = 4.25AEE27 pKa = 4.05ARR29 pKa = 11.84GDD31 pKa = 3.74GYY33 pKa = 11.72GDD35 pKa = 3.2IDD37 pKa = 4.02QISKK41 pKa = 10.49AGHH44 pKa = 6.35WDD46 pKa = 2.95QGTWGEE52 pKa = 3.93IDD54 pKa = 3.47AEE56 pKa = 4.15EE57 pKa = 4.89LIEE60 pKa = 5.37AGVDD64 pKa = 4.0FAKK67 pKa = 10.78LAVEE71 pKa = 4.52AGGEE75 pKa = 4.07SGRR78 pKa = 11.84AVDD81 pKa = 4.5LTLPTTPRR89 pKa = 11.84NLCGTACCFAGHH101 pKa = 6.45TALQVGDD108 pKa = 3.82RR109 pKa = 11.84PVVNVVVDD117 pKa = 3.45QALRR121 pKa = 11.84NDD123 pKa = 4.25LGSLTIMDD131 pKa = 4.49VRR133 pKa = 11.84PVDD136 pKa = 3.78NPDD139 pKa = 3.03VTVSVSTRR147 pKa = 11.84AMDD150 pKa = 4.3LLGLGHH156 pKa = 6.8EE157 pKa = 4.34EE158 pKa = 3.94ADD160 pKa = 4.12VLFEE164 pKa = 5.23ANNTLADD171 pKa = 4.02LRR173 pKa = 11.84VMVDD177 pKa = 3.52WICSGRR183 pKa = 11.84TVDD186 pKa = 3.9EE187 pKa = 5.21CPACGQRR194 pKa = 11.84PWDD197 pKa = 3.92CDD199 pKa = 3.37QSGEE203 pKa = 4.17ACEE206 pKa = 4.69DD207 pKa = 3.99CNEE210 pKa = 4.38HH211 pKa = 7.84LDD213 pKa = 4.74DD214 pKa = 5.67CEE216 pKa = 4.3CCADD220 pKa = 4.04CGAEE224 pKa = 3.94TGEE227 pKa = 4.32GCTCFTCMWCSSSTEE242 pKa = 4.29EE243 pKa = 5.45IDD245 pKa = 5.92GSDD248 pKa = 3.67CTCRR252 pKa = 11.84DD253 pKa = 3.95DD254 pKa = 5.79DD255 pKa = 5.99DD256 pKa = 4.1EE257 pKa = 6.93AEE259 pKa = 4.23GG260 pKa = 4.05
MM1 pKa = 7.38ATPTFEE7 pKa = 4.0PTTLRR12 pKa = 11.84GHH14 pKa = 6.5ALLRR18 pKa = 11.84HH19 pKa = 4.79VLEE22 pKa = 5.39IIEE25 pKa = 4.25AEE27 pKa = 4.05ARR29 pKa = 11.84GDD31 pKa = 3.74GYY33 pKa = 11.72GDD35 pKa = 3.2IDD37 pKa = 4.02QISKK41 pKa = 10.49AGHH44 pKa = 6.35WDD46 pKa = 2.95QGTWGEE52 pKa = 3.93IDD54 pKa = 3.47AEE56 pKa = 4.15EE57 pKa = 4.89LIEE60 pKa = 5.37AGVDD64 pKa = 4.0FAKK67 pKa = 10.78LAVEE71 pKa = 4.52AGGEE75 pKa = 4.07SGRR78 pKa = 11.84AVDD81 pKa = 4.5LTLPTTPRR89 pKa = 11.84NLCGTACCFAGHH101 pKa = 6.45TALQVGDD108 pKa = 3.82RR109 pKa = 11.84PVVNVVVDD117 pKa = 3.45QALRR121 pKa = 11.84NDD123 pKa = 4.25LGSLTIMDD131 pKa = 4.49VRR133 pKa = 11.84PVDD136 pKa = 3.78NPDD139 pKa = 3.03VTVSVSTRR147 pKa = 11.84AMDD150 pKa = 4.3LLGLGHH156 pKa = 6.8EE157 pKa = 4.34EE158 pKa = 3.94ADD160 pKa = 4.12VLFEE164 pKa = 5.23ANNTLADD171 pKa = 4.02LRR173 pKa = 11.84VMVDD177 pKa = 3.52WICSGRR183 pKa = 11.84TVDD186 pKa = 3.9EE187 pKa = 5.21CPACGQRR194 pKa = 11.84PWDD197 pKa = 3.92CDD199 pKa = 3.37QSGEE203 pKa = 4.17ACEE206 pKa = 4.69DD207 pKa = 3.99CNEE210 pKa = 4.38HH211 pKa = 7.84LDD213 pKa = 4.74DD214 pKa = 5.67CEE216 pKa = 4.3CCADD220 pKa = 4.04CGAEE224 pKa = 3.94TGEE227 pKa = 4.32GCTCFTCMWCSSSTEE242 pKa = 4.29EE243 pKa = 5.45IDD245 pKa = 5.92GSDD248 pKa = 3.67CTCRR252 pKa = 11.84DD253 pKa = 3.95DD254 pKa = 5.79DD255 pKa = 5.99DD256 pKa = 4.1EE257 pKa = 6.93AEE259 pKa = 4.23GG260 pKa = 4.05
Molecular weight: 27.84 kDa
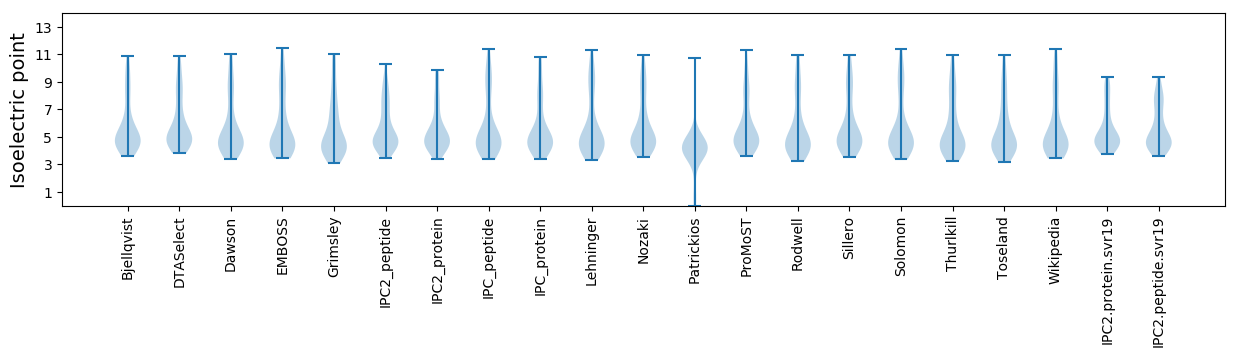
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J6TJY5|A0A5J6TJY5_9CAUD Uncharacterized protein OS=Gordonia phage CloverMinnie OX=2599842 GN=14 PE=4 SV=1
MM1 pKa = 7.57NKK3 pKa = 9.9RR4 pKa = 11.84LVYY7 pKa = 10.72VIGQPGSGKK16 pKa = 6.7TTLMSALTSASVRR29 pKa = 11.84LPQDD33 pKa = 3.0KK34 pKa = 9.8PVPRR38 pKa = 11.84DD39 pKa = 3.28LLATRR44 pKa = 11.84NTGRR48 pKa = 11.84VWAVEE53 pKa = 4.15LGRR56 pKa = 11.84QRR58 pKa = 11.84PGGFSGTDD66 pKa = 3.2ALASTAINTAEE77 pKa = 3.85PWLRR81 pKa = 11.84SQTEE85 pKa = 4.09APVVLAEE92 pKa = 4.18GARR95 pKa = 11.84LANRR99 pKa = 11.84RR100 pKa = 11.84FLTAAVEE107 pKa = 4.02SGYY110 pKa = 10.69QVLLVLLDD118 pKa = 3.73HH119 pKa = 7.45ADD121 pKa = 3.43AEE123 pKa = 4.52RR124 pKa = 11.84WRR126 pKa = 11.84EE127 pKa = 3.78ARR129 pKa = 11.84SVEE132 pKa = 4.22IGKK135 pKa = 8.63HH136 pKa = 5.13QNASWVKK143 pKa = 8.58GRR145 pKa = 11.84RR146 pKa = 11.84SASLNLAADD155 pKa = 4.13PPAGVEE161 pKa = 4.17VLRR164 pKa = 11.84GHH166 pKa = 7.18PSAVQEE172 pKa = 4.97PIWAWVRR179 pKa = 11.84ASS181 pKa = 3.14
MM1 pKa = 7.57NKK3 pKa = 9.9RR4 pKa = 11.84LVYY7 pKa = 10.72VIGQPGSGKK16 pKa = 6.7TTLMSALTSASVRR29 pKa = 11.84LPQDD33 pKa = 3.0KK34 pKa = 9.8PVPRR38 pKa = 11.84DD39 pKa = 3.28LLATRR44 pKa = 11.84NTGRR48 pKa = 11.84VWAVEE53 pKa = 4.15LGRR56 pKa = 11.84QRR58 pKa = 11.84PGGFSGTDD66 pKa = 3.2ALASTAINTAEE77 pKa = 3.85PWLRR81 pKa = 11.84SQTEE85 pKa = 4.09APVVLAEE92 pKa = 4.18GARR95 pKa = 11.84LANRR99 pKa = 11.84RR100 pKa = 11.84FLTAAVEE107 pKa = 4.02SGYY110 pKa = 10.69QVLLVLLDD118 pKa = 3.73HH119 pKa = 7.45ADD121 pKa = 3.43AEE123 pKa = 4.52RR124 pKa = 11.84WRR126 pKa = 11.84EE127 pKa = 3.78ARR129 pKa = 11.84SVEE132 pKa = 4.22IGKK135 pKa = 8.63HH136 pKa = 5.13QNASWVKK143 pKa = 8.58GRR145 pKa = 11.84RR146 pKa = 11.84SASLNLAADD155 pKa = 4.13PPAGVEE161 pKa = 4.17VLRR164 pKa = 11.84GHH166 pKa = 7.18PSAVQEE172 pKa = 4.97PIWAWVRR179 pKa = 11.84ASS181 pKa = 3.14
Molecular weight: 19.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
19696 |
55 |
1954 |
234.5 |
25.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.982 ± 0.277 | 0.99 ± 0.169 |
7.265 ± 0.296 | 6.159 ± 0.375 |
2.417 ± 0.169 | 8.398 ± 0.367 |
1.828 ± 0.154 | 4.509 ± 0.289 |
2.701 ± 0.224 | 8.22 ± 0.162 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.295 ± 0.121 | 2.777 ± 0.23 |
5.56 ± 0.238 | 3.437 ± 0.176 |
6.676 ± 0.39 | 5.788 ± 0.227 |
6.656 ± 0.195 | 7.773 ± 0.153 |
2.117 ± 0.122 | 2.452 ± 0.166 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
