
Kiloniella litopenaei
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Kiloniellaceae; Kiloniella
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
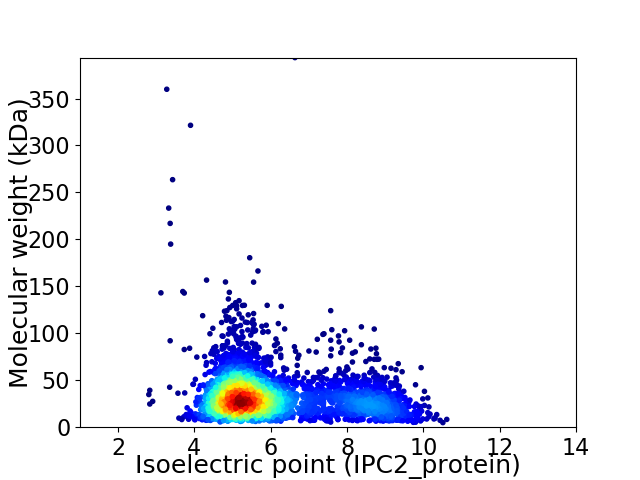
Virtual 2D-PAGE plot for 3735 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M2R471|A0A0M2R471_9PROT Bifunctional protein GlmU OS=Kiloniella litopenaei OX=1549748 GN=glmU PE=3 SV=1
MM1 pKa = 7.5KK2 pKa = 10.01KK3 pKa = 9.96ILMASTALAASLSYY17 pKa = 10.49AASASAEE24 pKa = 4.02DD25 pKa = 4.39TKK27 pKa = 11.07IKK29 pKa = 9.75WSAYY33 pKa = 7.57TDD35 pKa = 4.02FNVSSTSVDD44 pKa = 3.28GNVPGGDD51 pKa = 3.65TNNGLDD57 pKa = 3.6FNTNSEE63 pKa = 3.76IHH65 pKa = 6.5LNASQTADD73 pKa = 3.0NGLTYY78 pKa = 10.69GLHH81 pKa = 5.96IQLEE85 pKa = 4.34ADD87 pKa = 3.25QGNTNNTDD95 pKa = 3.23EE96 pKa = 4.04NHH98 pKa = 6.02IFIEE102 pKa = 4.53GDD104 pKa = 3.28FGRR107 pKa = 11.84VEE109 pKa = 5.76LGDD112 pKa = 3.44QDD114 pKa = 4.12SAGDD118 pKa = 3.34RR119 pKa = 11.84MMVSGSSVGFQYY131 pKa = 11.68GMFGNYY137 pKa = 8.64TGGLSALEE145 pKa = 4.34DD146 pKa = 3.67EE147 pKa = 5.24GNLGRR152 pKa = 11.84TAADD156 pKa = 3.98FSDD159 pKa = 4.1SSDD162 pKa = 3.4SSKK165 pKa = 9.26ITYY168 pKa = 7.58FTPTFNGFKK177 pKa = 10.21AGLSFAPDD185 pKa = 3.42SDD187 pKa = 3.74SGQTVSGNEE196 pKa = 3.68TDD198 pKa = 4.27FNNHH202 pKa = 5.33FDD204 pKa = 3.49AALNYY209 pKa = 10.63SGTFNDD215 pKa = 3.98FTVDD219 pKa = 3.16AALIGATHH227 pKa = 6.92EE228 pKa = 4.47VTSQGSSDD236 pKa = 3.37DD237 pKa = 3.11TYY239 pKa = 9.67YY240 pKa = 11.45AVGGGVMLGYY250 pKa = 10.81AGFKK254 pKa = 10.02AALGVLHH261 pKa = 7.74EE262 pKa = 4.45DD263 pKa = 3.43TKK265 pKa = 11.03DD266 pKa = 3.09TSEE269 pKa = 4.34INTVDD274 pKa = 2.9TGLSYY279 pKa = 11.3GIGAWSLSVGAAWSEE294 pKa = 3.62FDD296 pKa = 4.1QDD298 pKa = 3.34NGVEE302 pKa = 4.21NEE304 pKa = 4.35SIAYY308 pKa = 8.59SAGVGYY314 pKa = 10.18EE315 pKa = 3.88IAPGLTTWAGATIGDD330 pKa = 4.05YY331 pKa = 11.03EE332 pKa = 4.78GGVEE336 pKa = 5.42DD337 pKa = 4.15FTNFQTGISVAFF349 pKa = 4.0
MM1 pKa = 7.5KK2 pKa = 10.01KK3 pKa = 9.96ILMASTALAASLSYY17 pKa = 10.49AASASAEE24 pKa = 4.02DD25 pKa = 4.39TKK27 pKa = 11.07IKK29 pKa = 9.75WSAYY33 pKa = 7.57TDD35 pKa = 4.02FNVSSTSVDD44 pKa = 3.28GNVPGGDD51 pKa = 3.65TNNGLDD57 pKa = 3.6FNTNSEE63 pKa = 3.76IHH65 pKa = 6.5LNASQTADD73 pKa = 3.0NGLTYY78 pKa = 10.69GLHH81 pKa = 5.96IQLEE85 pKa = 4.34ADD87 pKa = 3.25QGNTNNTDD95 pKa = 3.23EE96 pKa = 4.04NHH98 pKa = 6.02IFIEE102 pKa = 4.53GDD104 pKa = 3.28FGRR107 pKa = 11.84VEE109 pKa = 5.76LGDD112 pKa = 3.44QDD114 pKa = 4.12SAGDD118 pKa = 3.34RR119 pKa = 11.84MMVSGSSVGFQYY131 pKa = 11.68GMFGNYY137 pKa = 8.64TGGLSALEE145 pKa = 4.34DD146 pKa = 3.67EE147 pKa = 5.24GNLGRR152 pKa = 11.84TAADD156 pKa = 3.98FSDD159 pKa = 4.1SSDD162 pKa = 3.4SSKK165 pKa = 9.26ITYY168 pKa = 7.58FTPTFNGFKK177 pKa = 10.21AGLSFAPDD185 pKa = 3.42SDD187 pKa = 3.74SGQTVSGNEE196 pKa = 3.68TDD198 pKa = 4.27FNNHH202 pKa = 5.33FDD204 pKa = 3.49AALNYY209 pKa = 10.63SGTFNDD215 pKa = 3.98FTVDD219 pKa = 3.16AALIGATHH227 pKa = 6.92EE228 pKa = 4.47VTSQGSSDD236 pKa = 3.37DD237 pKa = 3.11TYY239 pKa = 9.67YY240 pKa = 11.45AVGGGVMLGYY250 pKa = 10.81AGFKK254 pKa = 10.02AALGVLHH261 pKa = 7.74EE262 pKa = 4.45DD263 pKa = 3.43TKK265 pKa = 11.03DD266 pKa = 3.09TSEE269 pKa = 4.34INTVDD274 pKa = 2.9TGLSYY279 pKa = 11.3GIGAWSLSVGAAWSEE294 pKa = 3.62FDD296 pKa = 4.1QDD298 pKa = 3.34NGVEE302 pKa = 4.21NEE304 pKa = 4.35SIAYY308 pKa = 8.59SAGVGYY314 pKa = 10.18EE315 pKa = 3.88IAPGLTTWAGATIGDD330 pKa = 4.05YY331 pKa = 11.03EE332 pKa = 4.78GGVEE336 pKa = 5.42DD337 pKa = 4.15FTNFQTGISVAFF349 pKa = 4.0
Molecular weight: 36.49 kDa
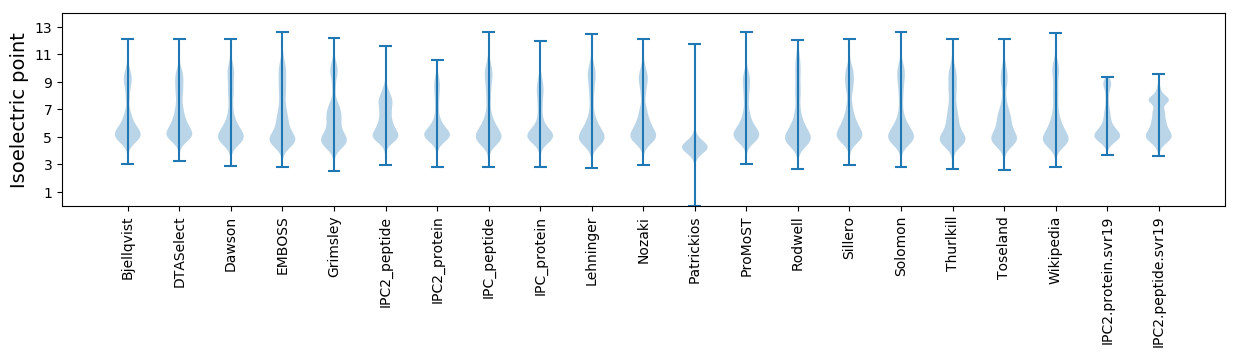
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M2R3M3|A0A0M2R3M3_9PROT Peptide transporter OS=Kiloniella litopenaei OX=1549748 GN=WH95_12990 PE=3 SV=1
MM1 pKa = 7.61AKK3 pKa = 9.99SFSSTLSVLFSLRR16 pKa = 11.84TVPVLTLMVFLLPVLAGLFSTIFPAFGYY44 pKa = 10.26LPAIGQVSFTTAAWTQLFAHH64 pKa = 7.14PALPNALLTSVSSGIIATLLSLILSLTFVAAWQGTNSFKK103 pKa = 10.81RR104 pKa = 11.84ARR106 pKa = 11.84QLLAPILAIPHH117 pKa = 5.66SALAIGLAFLIAPSGWITRR136 pKa = 11.84TISPWATGWEE146 pKa = 4.33RR147 pKa = 11.84PPDD150 pKa = 3.09IAVIHH155 pKa = 6.43DD156 pKa = 3.58EE157 pKa = 4.29WGLSLILGLMIKK169 pKa = 7.63EE170 pKa = 4.41TPYY173 pKa = 10.95LLLMIVAALDD183 pKa = 3.34QSQAKK188 pKa = 9.14QRR190 pKa = 11.84LKK192 pKa = 10.5LAKK195 pKa = 8.92TLGYY199 pKa = 9.99HH200 pKa = 6.61PVSAWIKK207 pKa = 9.2GVLPVIYY214 pKa = 9.83KK215 pKa = 9.38QIRR218 pKa = 11.84LPIYY222 pKa = 10.17AVLAFSLSVVDD233 pKa = 3.83IALILGPTTPPTLSILVLRR252 pKa = 11.84WFNDD256 pKa = 2.93PDD258 pKa = 3.37LSYY261 pKa = 10.33RR262 pKa = 11.84TIAAAGALLQLGVVVGVIFLWWATEE287 pKa = 4.0KK288 pKa = 10.82AVSRR292 pKa = 11.84LTRR295 pKa = 11.84RR296 pKa = 11.84WLANGKK302 pKa = 9.8RR303 pKa = 11.84SFAEE307 pKa = 3.92TPVRR311 pKa = 11.84IFSRR315 pKa = 11.84ISLNSIFVLFFGSLLVIVFWSFTRR339 pKa = 11.84RR340 pKa = 11.84WQYY343 pKa = 11.33PDD345 pKa = 4.08FLPSVWSIQSWDD357 pKa = 3.47RR358 pKa = 11.84NINNLLWSSSATLIIGLISSLIAVILVIACLEE390 pKa = 3.94NEE392 pKa = 3.92KK393 pKa = 10.38RR394 pKa = 11.84RR395 pKa = 11.84KK396 pKa = 9.7KK397 pKa = 10.58SAGTGALWLVYY408 pKa = 9.87LPLMIPQVSFLFGVQVLTIKK428 pKa = 10.84AGIDD432 pKa = 3.58ATWLAMIWGHH442 pKa = 6.54LLFVLPYY449 pKa = 10.27VFLSLTDD456 pKa = 4.04PYY458 pKa = 10.45RR459 pKa = 11.84QHH461 pKa = 7.49DD462 pKa = 3.6DD463 pKa = 3.53RR464 pKa = 11.84YY465 pKa = 9.82IKK467 pKa = 10.5VALSMGVSPTRR478 pKa = 11.84AFWRR482 pKa = 11.84IRR484 pKa = 11.84LPMLLRR490 pKa = 11.84PLLFALAIGFAVSVAQYY507 pKa = 10.99LPTLFIGGGRR517 pKa = 11.84FSTLTTEE524 pKa = 4.4AVNLSASGNRR534 pKa = 11.84RR535 pKa = 11.84IIAVYY540 pKa = 10.74ALLQALLPLLCFATALALPNVIFRR564 pKa = 11.84NRR566 pKa = 11.84RR567 pKa = 11.84TLRR570 pKa = 11.84EE571 pKa = 3.87HH572 pKa = 6.76
MM1 pKa = 7.61AKK3 pKa = 9.99SFSSTLSVLFSLRR16 pKa = 11.84TVPVLTLMVFLLPVLAGLFSTIFPAFGYY44 pKa = 10.26LPAIGQVSFTTAAWTQLFAHH64 pKa = 7.14PALPNALLTSVSSGIIATLLSLILSLTFVAAWQGTNSFKK103 pKa = 10.81RR104 pKa = 11.84ARR106 pKa = 11.84QLLAPILAIPHH117 pKa = 5.66SALAIGLAFLIAPSGWITRR136 pKa = 11.84TISPWATGWEE146 pKa = 4.33RR147 pKa = 11.84PPDD150 pKa = 3.09IAVIHH155 pKa = 6.43DD156 pKa = 3.58EE157 pKa = 4.29WGLSLILGLMIKK169 pKa = 7.63EE170 pKa = 4.41TPYY173 pKa = 10.95LLLMIVAALDD183 pKa = 3.34QSQAKK188 pKa = 9.14QRR190 pKa = 11.84LKK192 pKa = 10.5LAKK195 pKa = 8.92TLGYY199 pKa = 9.99HH200 pKa = 6.61PVSAWIKK207 pKa = 9.2GVLPVIYY214 pKa = 9.83KK215 pKa = 9.38QIRR218 pKa = 11.84LPIYY222 pKa = 10.17AVLAFSLSVVDD233 pKa = 3.83IALILGPTTPPTLSILVLRR252 pKa = 11.84WFNDD256 pKa = 2.93PDD258 pKa = 3.37LSYY261 pKa = 10.33RR262 pKa = 11.84TIAAAGALLQLGVVVGVIFLWWATEE287 pKa = 4.0KK288 pKa = 10.82AVSRR292 pKa = 11.84LTRR295 pKa = 11.84RR296 pKa = 11.84WLANGKK302 pKa = 9.8RR303 pKa = 11.84SFAEE307 pKa = 3.92TPVRR311 pKa = 11.84IFSRR315 pKa = 11.84ISLNSIFVLFFGSLLVIVFWSFTRR339 pKa = 11.84RR340 pKa = 11.84WQYY343 pKa = 11.33PDD345 pKa = 4.08FLPSVWSIQSWDD357 pKa = 3.47RR358 pKa = 11.84NINNLLWSSSATLIIGLISSLIAVILVIACLEE390 pKa = 3.94NEE392 pKa = 3.92KK393 pKa = 10.38RR394 pKa = 11.84RR395 pKa = 11.84KK396 pKa = 9.7KK397 pKa = 10.58SAGTGALWLVYY408 pKa = 9.87LPLMIPQVSFLFGVQVLTIKK428 pKa = 10.84AGIDD432 pKa = 3.58ATWLAMIWGHH442 pKa = 6.54LLFVLPYY449 pKa = 10.27VFLSLTDD456 pKa = 4.04PYY458 pKa = 10.45RR459 pKa = 11.84QHH461 pKa = 7.49DD462 pKa = 3.6DD463 pKa = 3.53RR464 pKa = 11.84YY465 pKa = 9.82IKK467 pKa = 10.5VALSMGVSPTRR478 pKa = 11.84AFWRR482 pKa = 11.84IRR484 pKa = 11.84LPMLLRR490 pKa = 11.84PLLFALAIGFAVSVAQYY507 pKa = 10.99LPTLFIGGGRR517 pKa = 11.84FSTLTTEE524 pKa = 4.4AVNLSASGNRR534 pKa = 11.84RR535 pKa = 11.84IIAVYY540 pKa = 10.74ALLQALLPLLCFATALALPNVIFRR564 pKa = 11.84NRR566 pKa = 11.84RR567 pKa = 11.84TLRR570 pKa = 11.84EE571 pKa = 3.87HH572 pKa = 6.76
Molecular weight: 63.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1159452 |
41 |
3542 |
310.4 |
34.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.074 ± 0.042 | 0.965 ± 0.013 |
6.046 ± 0.049 | 6.403 ± 0.043 |
4.052 ± 0.027 | 7.721 ± 0.047 |
2.047 ± 0.018 | 6.422 ± 0.034 |
5.214 ± 0.043 | 10.108 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.509 ± 0.021 | 3.785 ± 0.029 |
4.343 ± 0.025 | 3.537 ± 0.028 |
5.071 ± 0.039 | 6.412 ± 0.031 |
5.413 ± 0.032 | 6.973 ± 0.032 |
1.216 ± 0.018 | 2.687 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
