
Wenzhou sobemo-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
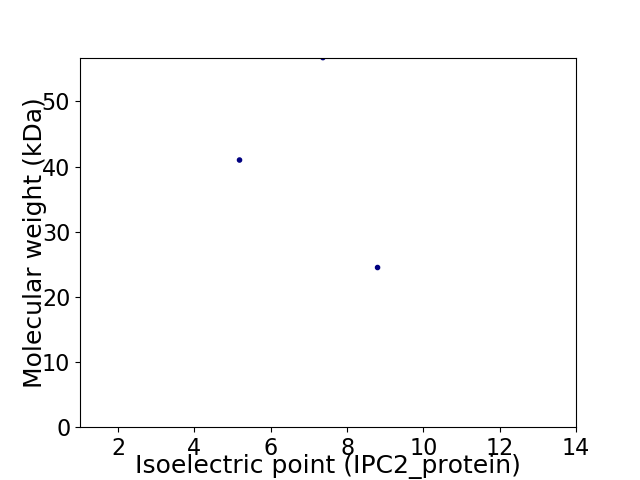
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
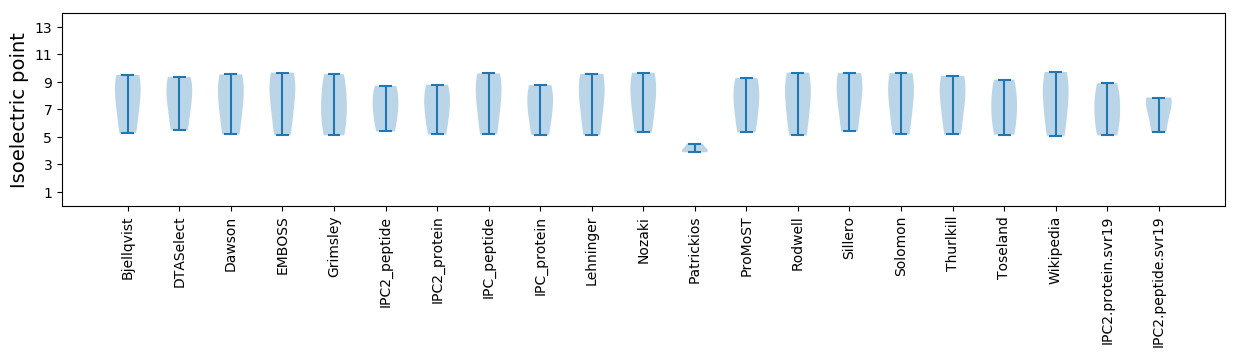
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEG5|A0A1L3KEG5_9VIRU Uncharacterized protein OS=Wenzhou sobemo-like virus 3 OX=1923659 PE=4 SV=1
MM1 pKa = 7.17FKK3 pKa = 10.61AAVMDD8 pKa = 4.72LDD10 pKa = 4.17MNSVPGQCEE19 pKa = 3.52MSRR22 pKa = 11.84FGSTNGEE29 pKa = 4.0ALGWDD34 pKa = 3.42ADD36 pKa = 3.99NYY38 pKa = 10.15SFDD41 pKa = 3.72YY42 pKa = 10.81EE43 pKa = 4.06RR44 pKa = 11.84YY45 pKa = 9.26HH46 pKa = 6.64YY47 pKa = 10.13FKK49 pKa = 10.89QLVKK53 pKa = 10.17TRR55 pKa = 11.84LLDD58 pKa = 3.71VIKK61 pKa = 10.79APVSDD66 pKa = 4.27EE67 pKa = 3.45IKK69 pKa = 10.37IFVKK73 pKa = 10.35QEE75 pKa = 3.56PHH77 pKa = 6.33KK78 pKa = 10.23KK79 pKa = 10.43AKK81 pKa = 9.91VDD83 pKa = 3.32EE84 pKa = 4.48GRR86 pKa = 11.84FRR88 pKa = 11.84LISAVSLVDD97 pKa = 3.62SMVDD101 pKa = 2.64RR102 pKa = 11.84MLFMRR107 pKa = 11.84ITHH110 pKa = 5.92RR111 pKa = 11.84VVPNYY116 pKa = 10.44VNTGVMIGWNPSAGGFRR133 pKa = 11.84YY134 pKa = 10.29LSALFPPGTPLLMADD149 pKa = 3.84RR150 pKa = 11.84SSWDD154 pKa = 2.91WTVQYY159 pKa = 11.76DD160 pKa = 3.72MIVAAKK166 pKa = 10.03EE167 pKa = 4.15VIQRR171 pKa = 11.84LAVGAPAWWSAAVEE185 pKa = 4.13HH186 pKa = 6.73RR187 pKa = 11.84FIALFEE193 pKa = 4.29NPVFRR198 pKa = 11.84FSDD201 pKa = 3.93GEE203 pKa = 4.17RR204 pKa = 11.84IMQQVPGIMKK214 pKa = 9.82SGCYY218 pKa = 8.46LTILLNSLIQLLLHH232 pKa = 6.42YY233 pKa = 9.5EE234 pKa = 4.1LSDD237 pKa = 3.51NPNIVVLGDD246 pKa = 3.5DD247 pKa = 4.04TLQEE251 pKa = 4.67DD252 pKa = 4.14EE253 pKa = 4.75DD254 pKa = 4.46DD255 pKa = 4.55EE256 pKa = 4.61YY257 pKa = 11.82LEE259 pKa = 4.15KK260 pKa = 11.01LRR262 pKa = 11.84DD263 pKa = 3.73FGITLKK269 pKa = 9.18VHH271 pKa = 5.72RR272 pKa = 11.84GEE274 pKa = 4.52RR275 pKa = 11.84IEE277 pKa = 4.07FAGFYY282 pKa = 10.17VDD284 pKa = 3.51EE285 pKa = 4.73KK286 pKa = 10.92GYY288 pKa = 10.77YY289 pKa = 8.16PAYY292 pKa = 9.08EE293 pKa = 4.5DD294 pKa = 3.11KK295 pKa = 11.09HH296 pKa = 7.02VFQLEE301 pKa = 4.37HH302 pKa = 6.17MCIDD306 pKa = 3.99DD307 pKa = 4.1PEE309 pKa = 5.23KK310 pKa = 11.06LGATLEE316 pKa = 4.25NYY318 pKa = 9.91QILYY322 pKa = 10.3ACVPAKK328 pKa = 10.68LKK330 pKa = 10.44VLRR333 pKa = 11.84SMVEE337 pKa = 3.73EE338 pKa = 3.9YY339 pKa = 11.16GLYY342 pKa = 10.49EE343 pKa = 4.22NYY345 pKa = 10.06ISDD348 pKa = 3.37GRR350 pKa = 11.84LRR352 pKa = 11.84GLKK355 pKa = 10.19HH356 pKa = 5.61GG357 pKa = 4.39
MM1 pKa = 7.17FKK3 pKa = 10.61AAVMDD8 pKa = 4.72LDD10 pKa = 4.17MNSVPGQCEE19 pKa = 3.52MSRR22 pKa = 11.84FGSTNGEE29 pKa = 4.0ALGWDD34 pKa = 3.42ADD36 pKa = 3.99NYY38 pKa = 10.15SFDD41 pKa = 3.72YY42 pKa = 10.81EE43 pKa = 4.06RR44 pKa = 11.84YY45 pKa = 9.26HH46 pKa = 6.64YY47 pKa = 10.13FKK49 pKa = 10.89QLVKK53 pKa = 10.17TRR55 pKa = 11.84LLDD58 pKa = 3.71VIKK61 pKa = 10.79APVSDD66 pKa = 4.27EE67 pKa = 3.45IKK69 pKa = 10.37IFVKK73 pKa = 10.35QEE75 pKa = 3.56PHH77 pKa = 6.33KK78 pKa = 10.23KK79 pKa = 10.43AKK81 pKa = 9.91VDD83 pKa = 3.32EE84 pKa = 4.48GRR86 pKa = 11.84FRR88 pKa = 11.84LISAVSLVDD97 pKa = 3.62SMVDD101 pKa = 2.64RR102 pKa = 11.84MLFMRR107 pKa = 11.84ITHH110 pKa = 5.92RR111 pKa = 11.84VVPNYY116 pKa = 10.44VNTGVMIGWNPSAGGFRR133 pKa = 11.84YY134 pKa = 10.29LSALFPPGTPLLMADD149 pKa = 3.84RR150 pKa = 11.84SSWDD154 pKa = 2.91WTVQYY159 pKa = 11.76DD160 pKa = 3.72MIVAAKK166 pKa = 10.03EE167 pKa = 4.15VIQRR171 pKa = 11.84LAVGAPAWWSAAVEE185 pKa = 4.13HH186 pKa = 6.73RR187 pKa = 11.84FIALFEE193 pKa = 4.29NPVFRR198 pKa = 11.84FSDD201 pKa = 3.93GEE203 pKa = 4.17RR204 pKa = 11.84IMQQVPGIMKK214 pKa = 9.82SGCYY218 pKa = 8.46LTILLNSLIQLLLHH232 pKa = 6.42YY233 pKa = 9.5EE234 pKa = 4.1LSDD237 pKa = 3.51NPNIVVLGDD246 pKa = 3.5DD247 pKa = 4.04TLQEE251 pKa = 4.67DD252 pKa = 4.14EE253 pKa = 4.75DD254 pKa = 4.46DD255 pKa = 4.55EE256 pKa = 4.61YY257 pKa = 11.82LEE259 pKa = 4.15KK260 pKa = 11.01LRR262 pKa = 11.84DD263 pKa = 3.73FGITLKK269 pKa = 9.18VHH271 pKa = 5.72RR272 pKa = 11.84GEE274 pKa = 4.52RR275 pKa = 11.84IEE277 pKa = 4.07FAGFYY282 pKa = 10.17VDD284 pKa = 3.51EE285 pKa = 4.73KK286 pKa = 10.92GYY288 pKa = 10.77YY289 pKa = 8.16PAYY292 pKa = 9.08EE293 pKa = 4.5DD294 pKa = 3.11KK295 pKa = 11.09HH296 pKa = 7.02VFQLEE301 pKa = 4.37HH302 pKa = 6.17MCIDD306 pKa = 3.99DD307 pKa = 4.1PEE309 pKa = 5.23KK310 pKa = 11.06LGATLEE316 pKa = 4.25NYY318 pKa = 9.91QILYY322 pKa = 10.3ACVPAKK328 pKa = 10.68LKK330 pKa = 10.44VLRR333 pKa = 11.84SMVEE337 pKa = 3.73EE338 pKa = 3.9YY339 pKa = 11.16GLYY342 pKa = 10.49EE343 pKa = 4.22NYY345 pKa = 10.06ISDD348 pKa = 3.37GRR350 pKa = 11.84LRR352 pKa = 11.84GLKK355 pKa = 10.19HH356 pKa = 5.61GG357 pKa = 4.39
Molecular weight: 41.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEG5|A0A1L3KEG5_9VIRU Uncharacterized protein OS=Wenzhou sobemo-like virus 3 OX=1923659 PE=4 SV=1
MM1 pKa = 7.66VSRR4 pKa = 11.84TDD6 pKa = 3.31TLEE9 pKa = 4.42AGLGTEE15 pKa = 4.78TIWSRR20 pKa = 11.84VFTLLTNTNVSKK32 pKa = 10.75SASFVARR39 pKa = 11.84EE40 pKa = 4.1KK41 pKa = 9.88VSKK44 pKa = 10.59YY45 pKa = 10.73YY46 pKa = 9.74EE47 pKa = 4.42TILCLGHH54 pKa = 7.14AISPGRR60 pKa = 11.84LLIHH64 pKa = 6.5VNGANWVFQQLNWCLPCLQRR84 pKa = 11.84SKK86 pKa = 10.8PQLTMVCNLLKK97 pKa = 10.57AVSRR101 pKa = 11.84SYY103 pKa = 11.12QYY105 pKa = 11.61LVTCLTQDD113 pKa = 3.06QHH115 pKa = 8.18GLGFRR120 pKa = 11.84VLHH123 pKa = 6.49IMLTTTYY130 pKa = 8.78MPCMSDD136 pKa = 3.17QVVLILEE143 pKa = 4.31LAVRR147 pKa = 11.84IYY149 pKa = 10.61MRR151 pKa = 11.84FCAWKK156 pKa = 9.24IRR158 pKa = 11.84RR159 pKa = 11.84TSSYY163 pKa = 8.58KK164 pKa = 9.67TSRR167 pKa = 11.84PLPANDD173 pKa = 3.15GSTRR177 pKa = 11.84VLAQLTLLNDD187 pKa = 3.55WWNTMAVCMFLIEE200 pKa = 5.32KK201 pKa = 9.5ILLMLSSILNGMTKK215 pKa = 10.6
MM1 pKa = 7.66VSRR4 pKa = 11.84TDD6 pKa = 3.31TLEE9 pKa = 4.42AGLGTEE15 pKa = 4.78TIWSRR20 pKa = 11.84VFTLLTNTNVSKK32 pKa = 10.75SASFVARR39 pKa = 11.84EE40 pKa = 4.1KK41 pKa = 9.88VSKK44 pKa = 10.59YY45 pKa = 10.73YY46 pKa = 9.74EE47 pKa = 4.42TILCLGHH54 pKa = 7.14AISPGRR60 pKa = 11.84LLIHH64 pKa = 6.5VNGANWVFQQLNWCLPCLQRR84 pKa = 11.84SKK86 pKa = 10.8PQLTMVCNLLKK97 pKa = 10.57AVSRR101 pKa = 11.84SYY103 pKa = 11.12QYY105 pKa = 11.61LVTCLTQDD113 pKa = 3.06QHH115 pKa = 8.18GLGFRR120 pKa = 11.84VLHH123 pKa = 6.49IMLTTTYY130 pKa = 8.78MPCMSDD136 pKa = 3.17QVVLILEE143 pKa = 4.31LAVRR147 pKa = 11.84IYY149 pKa = 10.61MRR151 pKa = 11.84FCAWKK156 pKa = 9.24IRR158 pKa = 11.84RR159 pKa = 11.84TSSYY163 pKa = 8.58KK164 pKa = 9.67TSRR167 pKa = 11.84PLPANDD173 pKa = 3.15GSTRR177 pKa = 11.84VLAQLTLLNDD187 pKa = 3.55WWNTMAVCMFLIEE200 pKa = 5.32KK201 pKa = 9.5ILLMLSSILNGMTKK215 pKa = 10.6
Molecular weight: 24.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1087 |
215 |
515 |
362.3 |
40.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.82 ± 1.221 | 1.472 ± 0.703 |
4.968 ± 1.148 | 5.244 ± 0.95 |
3.864 ± 0.453 | 5.98 ± 0.583 |
1.84 ± 0.312 | 4.14 ± 0.7 |
4.324 ± 0.443 | 9.936 ± 1.633 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.312 ± 0.582 | 3.588 ± 0.331 |
5.152 ± 0.987 | 3.68 ± 0.274 |
5.98 ± 0.167 | 7.82 ± 1.128 |
5.796 ± 1.499 | 8.464 ± 0.406 |
1.656 ± 0.373 | 4.968 ± 0.533 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
