
Hubei sobemo-like virus 46
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
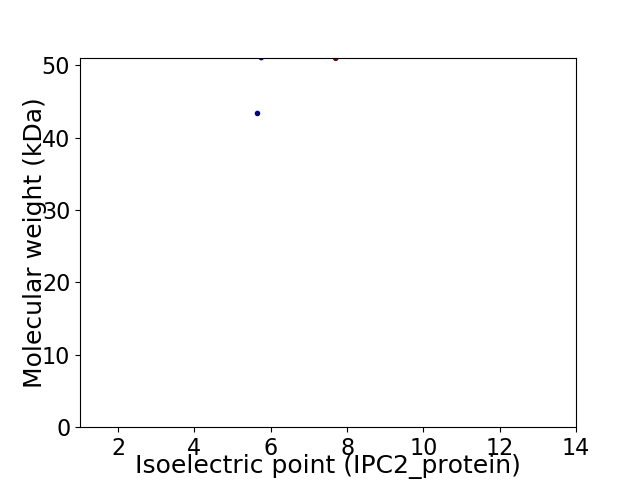
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEU4|A0A1L3KEU4_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 46 OX=1923234 PE=4 SV=1
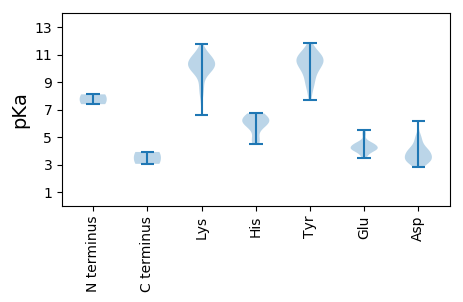
MM1 pKa = 7.39EE2 pKa = 4.1EE3 pKa = 4.47AYY5 pKa = 10.64RR6 pKa = 11.84EE7 pKa = 4.52VIWQLPDD14 pKa = 3.11NYY16 pKa = 10.79LSYY19 pKa = 10.96EE20 pKa = 4.0QFLVAVSRR28 pKa = 11.84VDD30 pKa = 3.51MKK32 pKa = 11.17SSPGWPYY39 pKa = 10.98VKK41 pKa = 10.37EE42 pKa = 4.22GSTNAQILQWNGFTCSEE59 pKa = 4.21YY60 pKa = 11.37ALGRR64 pKa = 11.84MWNEE68 pKa = 3.5VNHH71 pKa = 5.92YY72 pKa = 8.88LQSDD76 pKa = 3.99EE77 pKa = 5.48PIVLSCFIKK86 pKa = 10.62SEE88 pKa = 3.74PHH90 pKa = 6.68KK91 pKa = 10.29ISKK94 pKa = 8.52VQEE97 pKa = 4.11GRR99 pKa = 11.84WRR101 pKa = 11.84LIMASPLHH109 pKa = 5.88VQIVWNMFFMYY120 pKa = 10.64LNDD123 pKa = 3.89LMVKK127 pKa = 9.88KK128 pKa = 10.12AYY130 pKa = 9.94HH131 pKa = 6.37IPSQQGLVLPNGGWKK146 pKa = 10.13AYY148 pKa = 10.25LDD150 pKa = 3.4QWRR153 pKa = 11.84SKK155 pKa = 10.94GYY157 pKa = 10.5DD158 pKa = 3.03VGLDD162 pKa = 3.34KK163 pKa = 11.12QAWDD167 pKa = 3.17WTAPIWLISMCLEE180 pKa = 3.5LRR182 pKa = 11.84YY183 pKa = 9.67RR184 pKa = 11.84LCRR187 pKa = 11.84GRR189 pKa = 11.84MRR191 pKa = 11.84GQWYY195 pKa = 9.77NIARR199 pKa = 11.84KK200 pKa = 9.61LYY202 pKa = 9.73RR203 pKa = 11.84DD204 pKa = 3.61MYY206 pKa = 10.86DD207 pKa = 3.33NPTIMLSNGQVYY219 pKa = 9.04YY220 pKa = 8.39QTIPGIEE227 pKa = 4.0KK228 pKa = 9.95SGCVNTISDD237 pKa = 3.98NSFMQIGVHH246 pKa = 6.25VLVCLDD252 pKa = 4.64LGLPLYY258 pKa = 8.29PLPVAVGDD266 pKa = 4.45DD267 pKa = 3.89TLQCASQTADD277 pKa = 2.89VEE279 pKa = 4.66AYY281 pKa = 10.02AKK283 pKa = 10.48FGAIVKK289 pKa = 9.58SASAGIEE296 pKa = 4.09FVGHH300 pKa = 6.72EE301 pKa = 4.39FTTNGPVPLYY311 pKa = 10.38LGKK314 pKa = 9.94HH315 pKa = 4.84VVKK318 pKa = 10.65SMHH321 pKa = 6.12VADD324 pKa = 5.13EE325 pKa = 4.15YY326 pKa = 11.36LPEE329 pKa = 4.5YY330 pKa = 10.33LDD332 pKa = 3.42MMCRR336 pKa = 11.84MYY338 pKa = 10.66CKK340 pKa = 10.49SPIFSYY346 pKa = 9.32WACLANEE353 pKa = 4.42LNCPVFSQDD362 pKa = 4.11YY363 pKa = 7.72YY364 pKa = 11.24SRR366 pKa = 11.84WYY368 pKa = 10.22DD369 pKa = 3.39YY370 pKa = 11.13EE371 pKa = 4.42SRR373 pKa = 11.84FF374 pKa = 3.92
MM1 pKa = 7.39EE2 pKa = 4.1EE3 pKa = 4.47AYY5 pKa = 10.64RR6 pKa = 11.84EE7 pKa = 4.52VIWQLPDD14 pKa = 3.11NYY16 pKa = 10.79LSYY19 pKa = 10.96EE20 pKa = 4.0QFLVAVSRR28 pKa = 11.84VDD30 pKa = 3.51MKK32 pKa = 11.17SSPGWPYY39 pKa = 10.98VKK41 pKa = 10.37EE42 pKa = 4.22GSTNAQILQWNGFTCSEE59 pKa = 4.21YY60 pKa = 11.37ALGRR64 pKa = 11.84MWNEE68 pKa = 3.5VNHH71 pKa = 5.92YY72 pKa = 8.88LQSDD76 pKa = 3.99EE77 pKa = 5.48PIVLSCFIKK86 pKa = 10.62SEE88 pKa = 3.74PHH90 pKa = 6.68KK91 pKa = 10.29ISKK94 pKa = 8.52VQEE97 pKa = 4.11GRR99 pKa = 11.84WRR101 pKa = 11.84LIMASPLHH109 pKa = 5.88VQIVWNMFFMYY120 pKa = 10.64LNDD123 pKa = 3.89LMVKK127 pKa = 9.88KK128 pKa = 10.12AYY130 pKa = 9.94HH131 pKa = 6.37IPSQQGLVLPNGGWKK146 pKa = 10.13AYY148 pKa = 10.25LDD150 pKa = 3.4QWRR153 pKa = 11.84SKK155 pKa = 10.94GYY157 pKa = 10.5DD158 pKa = 3.03VGLDD162 pKa = 3.34KK163 pKa = 11.12QAWDD167 pKa = 3.17WTAPIWLISMCLEE180 pKa = 3.5LRR182 pKa = 11.84YY183 pKa = 9.67RR184 pKa = 11.84LCRR187 pKa = 11.84GRR189 pKa = 11.84MRR191 pKa = 11.84GQWYY195 pKa = 9.77NIARR199 pKa = 11.84KK200 pKa = 9.61LYY202 pKa = 9.73RR203 pKa = 11.84DD204 pKa = 3.61MYY206 pKa = 10.86DD207 pKa = 3.33NPTIMLSNGQVYY219 pKa = 9.04YY220 pKa = 8.39QTIPGIEE227 pKa = 4.0KK228 pKa = 9.95SGCVNTISDD237 pKa = 3.98NSFMQIGVHH246 pKa = 6.25VLVCLDD252 pKa = 4.64LGLPLYY258 pKa = 8.29PLPVAVGDD266 pKa = 4.45DD267 pKa = 3.89TLQCASQTADD277 pKa = 2.89VEE279 pKa = 4.66AYY281 pKa = 10.02AKK283 pKa = 10.48FGAIVKK289 pKa = 9.58SASAGIEE296 pKa = 4.09FVGHH300 pKa = 6.72EE301 pKa = 4.39FTTNGPVPLYY311 pKa = 10.38LGKK314 pKa = 9.94HH315 pKa = 4.84VVKK318 pKa = 10.65SMHH321 pKa = 6.12VADD324 pKa = 5.13EE325 pKa = 4.15YY326 pKa = 11.36LPEE329 pKa = 4.5YY330 pKa = 10.33LDD332 pKa = 3.42MMCRR336 pKa = 11.84MYY338 pKa = 10.66CKK340 pKa = 10.49SPIFSYY346 pKa = 9.32WACLANEE353 pKa = 4.42LNCPVFSQDD362 pKa = 4.11YY363 pKa = 7.72YY364 pKa = 11.24SRR366 pKa = 11.84WYY368 pKa = 10.22DD369 pKa = 3.39YY370 pKa = 11.13EE371 pKa = 4.42SRR373 pKa = 11.84FF374 pKa = 3.92
Molecular weight: 43.36 kDa
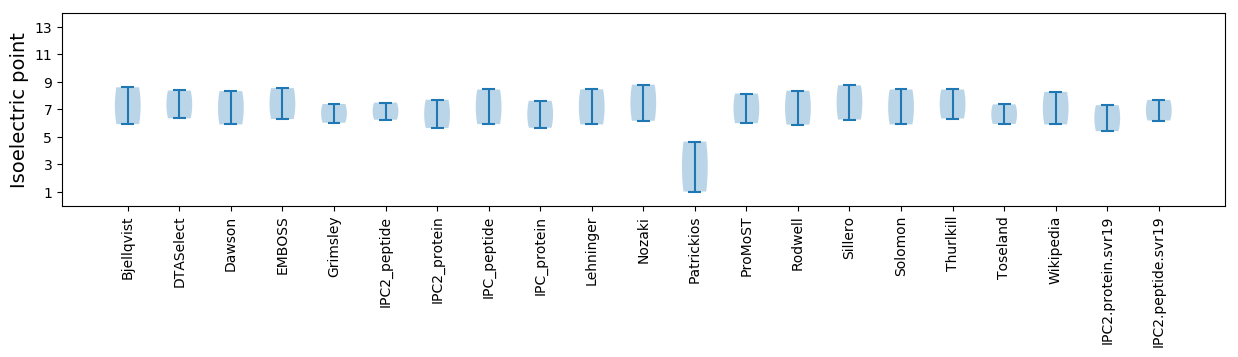
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEU4|A0A1L3KEU4_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 46 OX=1923234 PE=4 SV=1
MM1 pKa = 8.11DD2 pKa = 4.97KK3 pKa = 10.89FDD5 pKa = 6.14FEE7 pKa = 5.24ALMNRR12 pKa = 11.84IDD14 pKa = 4.04SLEE17 pKa = 3.95KK18 pKa = 10.74SIVPQSSGGILSHH31 pKa = 6.39LKK33 pKa = 9.74SWFEE37 pKa = 3.81NTVGRR42 pKa = 11.84FSRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 4.47IMYY50 pKa = 10.34AAGLAYY56 pKa = 10.08ALWLNRR62 pKa = 11.84RR63 pKa = 11.84LGPKK67 pKa = 10.03LGQLLRR73 pKa = 11.84WILSKK78 pKa = 10.71AVPGFRR84 pKa = 11.84QARR87 pKa = 11.84SWITGKK93 pKa = 10.2VEE95 pKa = 4.19ARR97 pKa = 11.84FEE99 pKa = 4.07PTRR102 pKa = 11.84LDD104 pKa = 3.14GRR106 pKa = 11.84EE107 pKa = 3.73NVMEE111 pKa = 4.57SIRR114 pKa = 11.84DD115 pKa = 3.8GSLLTPMMTPKK126 pKa = 10.39CQVSVGQMRR135 pKa = 11.84DD136 pKa = 3.43GEE138 pKa = 4.28FHH140 pKa = 6.04IHH142 pKa = 5.59GCGVRR147 pKa = 11.84LEE149 pKa = 4.45TFLVLPDD156 pKa = 4.26HH157 pKa = 6.22VWSYY161 pKa = 11.84SRR163 pKa = 11.84ISSEE167 pKa = 4.4TGRR170 pKa = 11.84TWLMGAAEE178 pKa = 4.31KK179 pKa = 10.21PIEE182 pKa = 4.12VSEE185 pKa = 4.68FEE187 pKa = 5.11ANVLDD192 pKa = 3.55TDD194 pKa = 3.79MVYY197 pKa = 10.71VRR199 pKa = 11.84LPNNVWSQLGVGTSALVHH217 pKa = 5.97EE218 pKa = 5.26VPSQGVYY225 pKa = 8.93CTIVGPATLGSCGMLRR241 pKa = 11.84HH242 pKa = 6.53DD243 pKa = 4.36PYY245 pKa = 10.76VFGRR249 pKa = 11.84VVYY252 pKa = 10.53DD253 pKa = 3.84GSTTGGFSGAAYY265 pKa = 8.95MRR267 pKa = 11.84ANSIAGIHH275 pKa = 4.8QRR277 pKa = 11.84GGKK280 pKa = 6.57TCNGGFSVSYY290 pKa = 10.44LWVTLCAMEE299 pKa = 4.38KK300 pKa = 9.59MKK302 pKa = 10.98PEE304 pKa = 4.24DD305 pKa = 3.32TADD308 pKa = 3.07WLRR311 pKa = 11.84NSFKK315 pKa = 10.15GKK317 pKa = 9.9KK318 pKa = 8.67RR319 pKa = 11.84VRR321 pKa = 11.84VDD323 pKa = 3.48KK324 pKa = 10.99SWADD328 pKa = 3.28TDD330 pKa = 4.02TVRR333 pKa = 11.84VMVDD337 pKa = 2.86GAFAIVEE344 pKa = 4.1RR345 pKa = 11.84SSMKK349 pKa = 10.1QAFGSNYY356 pKa = 9.58QDD358 pKa = 3.88DD359 pKa = 5.03LEE361 pKa = 4.58DD362 pKa = 4.0FKK364 pKa = 11.74VPRR367 pKa = 11.84EE368 pKa = 4.1LGYY371 pKa = 10.95EE372 pKa = 4.02DD373 pKa = 4.67AVNEE377 pKa = 4.18SGEE380 pKa = 4.43VKK382 pKa = 10.47SLKK385 pKa = 10.25QPGALSVVQNTQEE398 pKa = 4.32EE399 pKa = 4.68EE400 pKa = 4.15TSPVLILTNGFKK412 pKa = 10.57RR413 pKa = 11.84LSTEE417 pKa = 3.67QQNMVMGEE425 pKa = 3.95LNALRR430 pKa = 11.84KK431 pKa = 7.81ITNIRR436 pKa = 11.84KK437 pKa = 8.2MALQSKK443 pKa = 8.94AASSSIVTDD452 pKa = 3.5SSSTQQAA459 pKa = 3.05
MM1 pKa = 8.11DD2 pKa = 4.97KK3 pKa = 10.89FDD5 pKa = 6.14FEE7 pKa = 5.24ALMNRR12 pKa = 11.84IDD14 pKa = 4.04SLEE17 pKa = 3.95KK18 pKa = 10.74SIVPQSSGGILSHH31 pKa = 6.39LKK33 pKa = 9.74SWFEE37 pKa = 3.81NTVGRR42 pKa = 11.84FSRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 4.47IMYY50 pKa = 10.34AAGLAYY56 pKa = 10.08ALWLNRR62 pKa = 11.84RR63 pKa = 11.84LGPKK67 pKa = 10.03LGQLLRR73 pKa = 11.84WILSKK78 pKa = 10.71AVPGFRR84 pKa = 11.84QARR87 pKa = 11.84SWITGKK93 pKa = 10.2VEE95 pKa = 4.19ARR97 pKa = 11.84FEE99 pKa = 4.07PTRR102 pKa = 11.84LDD104 pKa = 3.14GRR106 pKa = 11.84EE107 pKa = 3.73NVMEE111 pKa = 4.57SIRR114 pKa = 11.84DD115 pKa = 3.8GSLLTPMMTPKK126 pKa = 10.39CQVSVGQMRR135 pKa = 11.84DD136 pKa = 3.43GEE138 pKa = 4.28FHH140 pKa = 6.04IHH142 pKa = 5.59GCGVRR147 pKa = 11.84LEE149 pKa = 4.45TFLVLPDD156 pKa = 4.26HH157 pKa = 6.22VWSYY161 pKa = 11.84SRR163 pKa = 11.84ISSEE167 pKa = 4.4TGRR170 pKa = 11.84TWLMGAAEE178 pKa = 4.31KK179 pKa = 10.21PIEE182 pKa = 4.12VSEE185 pKa = 4.68FEE187 pKa = 5.11ANVLDD192 pKa = 3.55TDD194 pKa = 3.79MVYY197 pKa = 10.71VRR199 pKa = 11.84LPNNVWSQLGVGTSALVHH217 pKa = 5.97EE218 pKa = 5.26VPSQGVYY225 pKa = 8.93CTIVGPATLGSCGMLRR241 pKa = 11.84HH242 pKa = 6.53DD243 pKa = 4.36PYY245 pKa = 10.76VFGRR249 pKa = 11.84VVYY252 pKa = 10.53DD253 pKa = 3.84GSTTGGFSGAAYY265 pKa = 8.95MRR267 pKa = 11.84ANSIAGIHH275 pKa = 4.8QRR277 pKa = 11.84GGKK280 pKa = 6.57TCNGGFSVSYY290 pKa = 10.44LWVTLCAMEE299 pKa = 4.38KK300 pKa = 9.59MKK302 pKa = 10.98PEE304 pKa = 4.24DD305 pKa = 3.32TADD308 pKa = 3.07WLRR311 pKa = 11.84NSFKK315 pKa = 10.15GKK317 pKa = 9.9KK318 pKa = 8.67RR319 pKa = 11.84VRR321 pKa = 11.84VDD323 pKa = 3.48KK324 pKa = 10.99SWADD328 pKa = 3.28TDD330 pKa = 4.02TVRR333 pKa = 11.84VMVDD337 pKa = 2.86GAFAIVEE344 pKa = 4.1RR345 pKa = 11.84SSMKK349 pKa = 10.1QAFGSNYY356 pKa = 9.58QDD358 pKa = 3.88DD359 pKa = 5.03LEE361 pKa = 4.58DD362 pKa = 4.0FKK364 pKa = 11.74VPRR367 pKa = 11.84EE368 pKa = 4.1LGYY371 pKa = 10.95EE372 pKa = 4.02DD373 pKa = 4.67AVNEE377 pKa = 4.18SGEE380 pKa = 4.43VKK382 pKa = 10.47SLKK385 pKa = 10.25QPGALSVVQNTQEE398 pKa = 4.32EE399 pKa = 4.68EE400 pKa = 4.15TSPVLILTNGFKK412 pKa = 10.57RR413 pKa = 11.84LSTEE417 pKa = 3.67QQNMVMGEE425 pKa = 3.95LNALRR430 pKa = 11.84KK431 pKa = 7.81ITNIRR436 pKa = 11.84KK437 pKa = 8.2MALQSKK443 pKa = 8.94AASSSIVTDD452 pKa = 3.5SSSTQQAA459 pKa = 3.05
Molecular weight: 50.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
833 |
374 |
459 |
416.5 |
47.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.122 ± 0.309 | 2.041 ± 0.549 |
4.922 ± 0.096 | 5.642 ± 0.343 |
3.481 ± 0.166 | 7.563 ± 0.862 |
1.921 ± 0.133 | 4.442 ± 0.389 |
4.802 ± 0.156 | 8.643 ± 0.273 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.082 ± 0.12 | 3.962 ± 0.193 |
4.202 ± 0.536 | 4.322 ± 0.299 |
5.642 ± 0.832 | 8.403 ± 0.722 |
4.322 ± 1.005 | 8.043 ± 0.339 |
2.881 ± 0.526 | 4.562 ± 1.62 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
