
Synchytrium endobioticum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Chytridiomycota; Chytridiomycota incertae sedis; Chytridiomycetes; Synchytriales; Synchytriaceae; Synchytrium
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
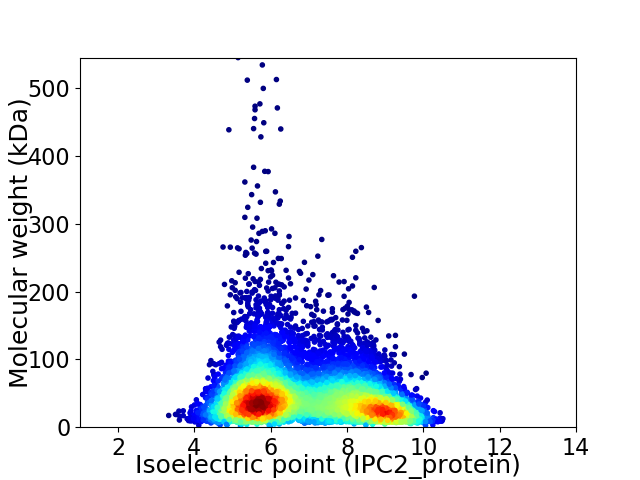
Virtual 2D-PAGE plot for 7895 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
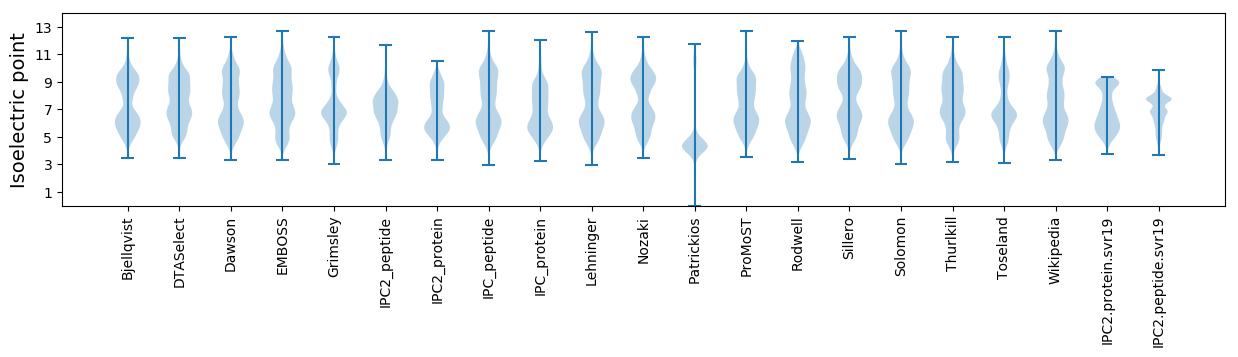
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A507DGB9|A0A507DGB9_9FUNG WD_REPEATS_REGION domain-containing protein OS=Synchytrium endobioticum OX=286115 GN=SeLEV6574_g01199 PE=4 SV=1
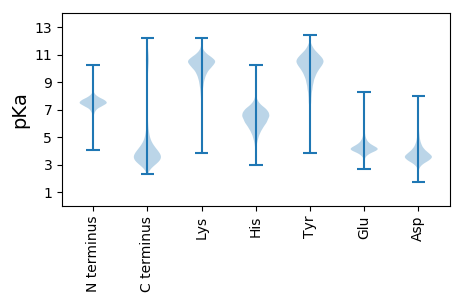
MM1 pKa = 7.15LTLRR5 pKa = 11.84QTVVEE10 pKa = 4.37LIIMVMLMSSNVLLSPVPMGACFSSTRR37 pKa = 11.84GSAEE41 pKa = 3.85VVAPDD46 pKa = 3.59YY47 pKa = 11.33PNDD50 pKa = 3.27VVAAHH55 pKa = 5.68YY56 pKa = 10.52HH57 pKa = 5.61NNDD60 pKa = 3.27DD61 pKa = 4.32RR62 pKa = 11.84TDD64 pKa = 3.39SSDD67 pKa = 3.57DD68 pKa = 3.45LRR70 pKa = 11.84GVNIPFSDD78 pKa = 4.02GEE80 pKa = 4.23TDD82 pKa = 3.49GPEE85 pKa = 4.9DD86 pKa = 3.84YY87 pKa = 10.4FSPSSDD93 pKa = 2.85PVVSTEE99 pKa = 3.98VSSDD103 pKa = 3.16VCMTICQSRR112 pKa = 11.84STSTVTGSVSLVV124 pKa = 3.14
MM1 pKa = 7.15LTLRR5 pKa = 11.84QTVVEE10 pKa = 4.37LIIMVMLMSSNVLLSPVPMGACFSSTRR37 pKa = 11.84GSAEE41 pKa = 3.85VVAPDD46 pKa = 3.59YY47 pKa = 11.33PNDD50 pKa = 3.27VVAAHH55 pKa = 5.68YY56 pKa = 10.52HH57 pKa = 5.61NNDD60 pKa = 3.27DD61 pKa = 4.32RR62 pKa = 11.84TDD64 pKa = 3.39SSDD67 pKa = 3.57DD68 pKa = 3.45LRR70 pKa = 11.84GVNIPFSDD78 pKa = 4.02GEE80 pKa = 4.23TDD82 pKa = 3.49GPEE85 pKa = 4.9DD86 pKa = 3.84YY87 pKa = 10.4FSPSSDD93 pKa = 2.85PVVSTEE99 pKa = 3.98VSSDD103 pKa = 3.16VCMTICQSRR112 pKa = 11.84STSTVTGSVSLVV124 pKa = 3.14
Molecular weight: 13.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A507DSE5|A0A507DSE5_9FUNG RRM domain-containing protein OS=Synchytrium endobioticum OX=286115 GN=SeMB42_g00139 PE=3 SV=1
MM1 pKa = 7.47LRR3 pKa = 11.84AQHH6 pKa = 6.33EE7 pKa = 4.51PTVQRR12 pKa = 11.84SPTISIITPKK22 pKa = 9.73MLSEE26 pKa = 3.93RR27 pKa = 11.84ATLNRR32 pKa = 11.84HH33 pKa = 4.71PAKK36 pKa = 10.16PKK38 pKa = 10.55APTVPLSRR46 pKa = 11.84TPARR50 pKa = 11.84TPVALSASTSISSTRR65 pKa = 11.84AAPARR70 pKa = 11.84SAPARR75 pKa = 11.84SEE77 pKa = 3.76NCATSKK83 pKa = 11.14ASFASTPRR91 pKa = 11.84FSSAVTGRR99 pKa = 11.84EE100 pKa = 3.65RR101 pKa = 11.84LTGKK105 pKa = 9.53HH106 pKa = 5.78APAATLRR113 pKa = 11.84AAIQEE118 pKa = 4.01PFMGKK123 pKa = 8.23NHH125 pKa = 5.53STAFGTVYY133 pKa = 10.05TRR135 pKa = 11.84GGIPCRR141 pKa = 11.84LQHH144 pKa = 6.73GSVQHH149 pKa = 6.71RR150 pKa = 11.84IQWTTSPPSLSFPTLLPLFIVGLLEE175 pKa = 3.55THH177 pKa = 5.87PQYY180 pKa = 11.23RR181 pKa = 11.84FIAHH185 pKa = 6.06QGLMEE190 pKa = 5.59CISAPNCGDD199 pKa = 4.38SIQQAWTVAVPPLRR213 pKa = 11.84GVLGNSGDD221 pKa = 3.57KK222 pKa = 10.98GALLTALKK230 pKa = 10.1ALAAMIPNLSIDD242 pKa = 4.18SLLSACGMLLPPLSRR257 pKa = 11.84LAVSKK262 pKa = 10.83DD263 pKa = 3.24RR264 pKa = 11.84EE265 pKa = 4.52VNDD268 pKa = 3.79ATHH271 pKa = 6.47SMLRR275 pKa = 11.84TLEE278 pKa = 3.98EE279 pKa = 3.77RR280 pKa = 11.84ALEE283 pKa = 4.02YY284 pKa = 10.38PQVLKK289 pKa = 10.79LIKK292 pKa = 10.36ARR294 pKa = 11.84IPTYY298 pKa = 10.31TSIRR302 pKa = 11.84SCC304 pKa = 3.89
MM1 pKa = 7.47LRR3 pKa = 11.84AQHH6 pKa = 6.33EE7 pKa = 4.51PTVQRR12 pKa = 11.84SPTISIITPKK22 pKa = 9.73MLSEE26 pKa = 3.93RR27 pKa = 11.84ATLNRR32 pKa = 11.84HH33 pKa = 4.71PAKK36 pKa = 10.16PKK38 pKa = 10.55APTVPLSRR46 pKa = 11.84TPARR50 pKa = 11.84TPVALSASTSISSTRR65 pKa = 11.84AAPARR70 pKa = 11.84SAPARR75 pKa = 11.84SEE77 pKa = 3.76NCATSKK83 pKa = 11.14ASFASTPRR91 pKa = 11.84FSSAVTGRR99 pKa = 11.84EE100 pKa = 3.65RR101 pKa = 11.84LTGKK105 pKa = 9.53HH106 pKa = 5.78APAATLRR113 pKa = 11.84AAIQEE118 pKa = 4.01PFMGKK123 pKa = 8.23NHH125 pKa = 5.53STAFGTVYY133 pKa = 10.05TRR135 pKa = 11.84GGIPCRR141 pKa = 11.84LQHH144 pKa = 6.73GSVQHH149 pKa = 6.71RR150 pKa = 11.84IQWTTSPPSLSFPTLLPLFIVGLLEE175 pKa = 3.55THH177 pKa = 5.87PQYY180 pKa = 11.23RR181 pKa = 11.84FIAHH185 pKa = 6.06QGLMEE190 pKa = 5.59CISAPNCGDD199 pKa = 4.38SIQQAWTVAVPPLRR213 pKa = 11.84GVLGNSGDD221 pKa = 3.57KK222 pKa = 10.98GALLTALKK230 pKa = 10.1ALAAMIPNLSIDD242 pKa = 4.18SLLSACGMLLPPLSRR257 pKa = 11.84LAVSKK262 pKa = 10.83DD263 pKa = 3.24RR264 pKa = 11.84EE265 pKa = 4.52VNDD268 pKa = 3.79ATHH271 pKa = 6.47SMLRR275 pKa = 11.84TLEE278 pKa = 3.98EE279 pKa = 3.77RR280 pKa = 11.84ALEE283 pKa = 4.02YY284 pKa = 10.38PQVLKK289 pKa = 10.79LIKK292 pKa = 10.36ARR294 pKa = 11.84IPTYY298 pKa = 10.31TSIRR302 pKa = 11.84SCC304 pKa = 3.89
Molecular weight: 32.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3746747 |
8 |
4874 |
474.6 |
52.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.44 ± 0.028 | 1.393 ± 0.011 |
5.793 ± 0.018 | 5.644 ± 0.027 |
3.382 ± 0.014 | 5.737 ± 0.021 |
2.729 ± 0.013 | 5.205 ± 0.016 |
5.153 ± 0.023 | 9.192 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.385 ± 0.01 | 4.019 ± 0.015 |
5.774 ± 0.027 | 3.972 ± 0.019 |
6.02 ± 0.02 | 8.705 ± 0.032 |
6.081 ± 0.019 | 6.409 ± 0.018 |
1.144 ± 0.008 | 2.817 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
